Gene Page: EPHA5
Summary ?
| GeneID | 2044 |
| Symbol | EPHA5 |
| Synonyms | CEK7|EHK-1|EHK1|EK7|HEK7|TYRO4 |
| Description | EPH receptor A5 |
| Reference | MIM:600004|HGNC:HGNC:3389|Ensembl:ENSG00000145242|HPRD:02474|Vega:OTTHUMG00000129273 |
| Gene type | protein-coding |
| Map location | 4q13.1 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 0.364 |
| Fetal beta | 1.96 |
| eGene | Cerebellar Hemisphere Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs4404602 | 4 | 0 | null | 0.4229 | EPHA5 | null |
Section II. Transcriptome annotation
General gene expression (GTEx)
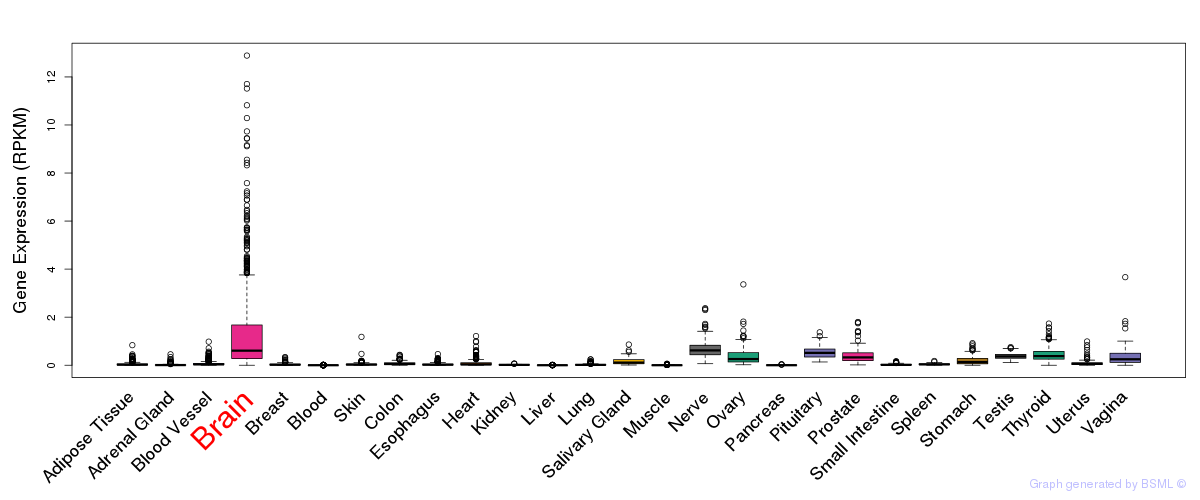
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBCCD1 | 0.83 | 0.77 |
| L3MBTL3 | 0.82 | 0.76 |
| SLC44A5 | 0.82 | 0.76 |
| FAM126A | 0.82 | 0.82 |
| NCUBE1 | 0.82 | 0.79 |
| FCHO2 | 0.82 | 0.82 |
| SCN3A | 0.82 | 0.78 |
| SATB2 | 0.81 | 0.63 |
| PTPRO | 0.81 | 0.76 |
| NEO1 | 0.81 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.62 | -0.64 |
| CA4 | -0.60 | -0.67 |
| HLA-F | -0.59 | -0.62 |
| MT-CO2 | -0.59 | -0.68 |
| FXYD1 | -0.58 | -0.65 |
| IFI27 | -0.57 | -0.65 |
| AF347015.27 | -0.57 | -0.64 |
| AF347015.31 | -0.57 | -0.66 |
| MYL3 | -0.57 | -0.66 |
| AF347015.33 | -0.56 | -0.65 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PIEPOLI LGI1 TARGETS UP | 15 | 11 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA LOH REGIONS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| NOUZOVA METHYLATED IN APL | 68 | 39 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM UP | 38 | 30 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |