Gene Page: ERBB3
Summary ?
| GeneID | 2065 |
| Symbol | ERBB3 |
| Synonyms | ErbB-3|HER3|LCCS2|MDA-BF-1|c-erbB-3|c-erbB3|erbB3-S|p180-ErbB3|p45-sErbB3|p85-sErbB3 |
| Description | erb-b2 receptor tyrosine kinase 3 |
| Reference | MIM:190151|HGNC:HGNC:3431|Ensembl:ENSG00000065361|HPRD:01820|Vega:OTTHUMG00000170140 |
| Gene type | protein-coding |
| Map location | 12q13 |
| Pascal p-value | 0.374 |
| Sherlock p-value | 0.068 |
| Fetal beta | -2.664 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 7.3266 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs973234 | chr3 | 189840350 | ERBB3 | 2065 | 0.14 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
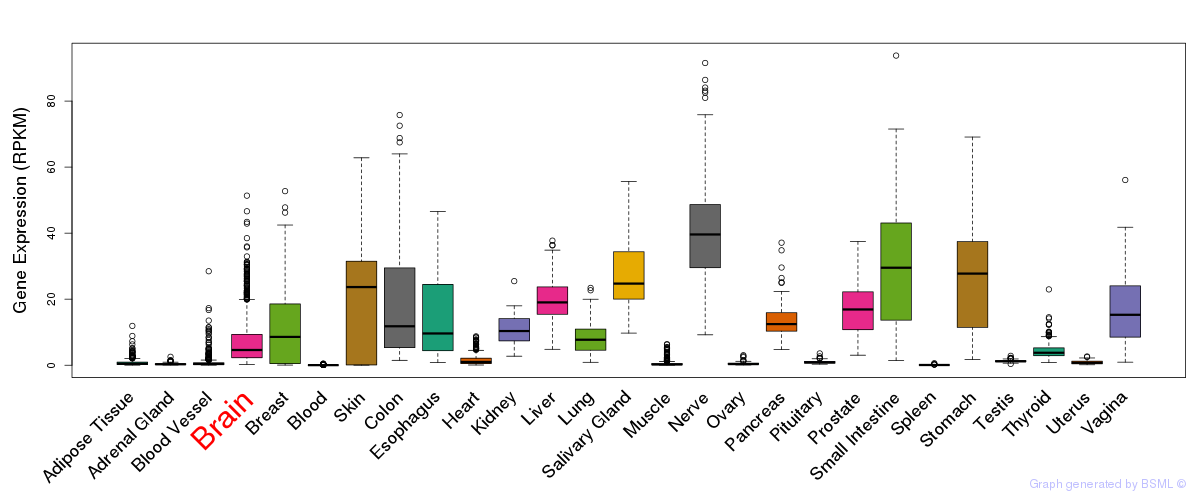
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | IEA | neurite (GO term level: 8) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004888 | transmembrane receptor activity | ISS | 7514177 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IGI | 7556068 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0019838 | growth factor binding | IPI | 11389077 | |
| GO:0019838 | growth factor binding | ISS | 7514177 | |
| GO:0042803 | protein homodimerization activity | NAS | 11389077 | |
| GO:0030296 | protein tyrosine kinase activator activity | IDA | 7556068 | |
| GO:0046982 | protein heterodimerization activity | IDA | 10572067 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0014037 | Schwann cell differentiation | IEA | neuron, axon, Glial (GO term level: 9) | - |
| GO:0021545 | cranial nerve development | IEA | Brain (GO term level: 7) | - |
| GO:0043524 | negative regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IEA | - | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IGI | 7556068 | |
| GO:0007162 | negative regulation of cell adhesion | IDA | 7556068 | |
| GO:0009968 | negative regulation of signal transduction | IDA | 11389077 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0014068 | positive regulation of phosphoinositide 3-kinase cascade | TAS | 17701904 | |
| GO:0042060 | wound healing | NAS | 12646923 | |
| GO:0051048 | negative regulation of secretion | IDA | 10559227 | |
| GO:0042127 | regulation of cell proliferation | IDA | 11389077 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043235 | receptor complex | ISS | Neurotransmitter (GO term level: 4) | 7514177 |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 11389077 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 2687875 | |
| GO:0016323 | basolateral plasma membrane | IDA | 12646923 | |
| GO:0016324 | apical plasma membrane | IEA | - | |
| GO:0016328 | lateral plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AGTR2 | AT2 | ATGR2 | MRX88 | angiotensin II receptor, type 2 | - | HPRD | 10710290 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD,BioGRID | 14576349 |
| CSK | MGC117393 | c-src tyrosine kinase | Affinity Capture-MS | BioGRID | 16729043 |
| EGF | HOMG4 | URG | epidermal growth factor (beta-urogastrone) | - | HPRD,BioGRID | 12093292 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 10527633 |
| EGR1 | AT225 | G0S30 | KROX-24 | NGFI-A | TIS8 | ZIF-268 | ZNF225 | early growth response 1 | Two-hybrid | BioGRID | 12840049 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 7538656 |10799311 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 7538656 |
| ERBB3 | ErbB-3 | HER3 | LCCS2 | MDA-BF-1 | MGC88033 | c-erbB-3 | c-erbB3 | erbB3-S | p180-ErbB3 | p45-sErbB3 | p85-sErbB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | - | HPRD | 12154198 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-MS | BioGRID | 16729043 |
| GRB7 | - | growth factor receptor-bound protein 7 | - | HPRD,BioGRID | 9516479 |
| IL6ST | CD130 | CDw130 | GP130 | GP130-RAPS | IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | - | HPRD,BioGRID | 11821958 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | - | HPRD,BioGRID | 11278868 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | Reconstituted Complex | BioGRID | 7592681 |9168115 |11555649 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | NDF-alpha interacts with Her3. An interaction between human NDF-alpha and Her3 from an unknown species was demonstrated by experimental system. | BIND | 7592681 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | - | HPRD | 7592681 |11555649 |
| NRG2 | Don-1 | HRG2 | NTAK | neuregulin 2 | - | HPRD,BioGRID | 9168114 |9168115 |
| ODF2L | KIAA1229 | MGC111060 | RP5-977L11.1 | dJ977L11.1 | outer dense fiber of sperm tails 2-like | Two-hybrid | BioGRID | 16169070 |
| PA2G4 | EBP1 | HG4-1 | p38-2G4 | proliferation-associated 2G4, 38kDa | ErbB-3 interacts with EBP1. | BIND | 10682683 |
| PA2G4 | EBP1 | HG4-1 | p38-2G4 | proliferation-associated 2G4, 38kDa | - | HPRD,BioGRID | 10682683 |11325528 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 10383151 |11546794 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | Affinity Capture-MS | BioGRID | 16729043 |
| PTGES3 | P23 | TEBP | cPGES | prostaglandin E synthase 3 (cytosolic) | - | HPRD | 10095121 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 11807823 |
| PTK6 | BRK | FLJ42088 | PTK6 protein tyrosine kinase 6 | - | HPRD,BioGRID | 11114724 |
| RGS4 | DKFZp761F1924 | MGC2124 | MGC60244 | RGP4 | SCZD9 | regulator of G-protein signaling 4 | - | HPRD,BioGRID | 12840049 |
| RNF41 | MGC45228 | NRDP1 | SBBI03 | ring finger protein 41 | Reconstituted Complex | BioGRID | 11867753 |
| RPN1 | DKFZp686B16177 | OST1 | RBPH1 | ribophorin I | Two-hybrid | BioGRID | 12840049 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD | 9694850 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Affinity Capture-MS | BioGRID | 16729043 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | - | HPRD | 11173924 |
| ZNF207 | DKFZp761N202 | zinc finger protein 207 | Two-hybrid | BioGRID | 12840049 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HER2 PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID ERBB NETWORK PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| LUCAS HNF4A TARGETS UP | 58 | 36 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP | 81 | 57 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 1 | 13 | 10 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS UP | 92 | 64 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| JEON SMAD6 TARGETS DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL DN | 82 | 52 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| WHITESIDE CISPLATIN RESISTANCE UP | 11 | 7 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C2 | 54 | 39 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCERS KINOME BLUE | 21 | 16 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| WU SILENCED BY METHYLATION IN BLADDER CANCER | 55 | 42 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP | 68 | 49 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-143 | 1130 | 1137 | 1A,m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA | ||||
| miR-148/152 | 1077 | 1084 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-205 | 287 | 294 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-22 | 36 | 43 | 1A,m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-378 | 1135 | 1141 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.