Gene Page: ERBB4
Summary ?
| GeneID | 2066 |
| Symbol | ERBB4 |
| Synonyms | ALS19|HER4|p180erbB4 |
| Description | erb-b2 receptor tyrosine kinase 4 |
| Reference | MIM:600543|HGNC:HGNC:3432|Ensembl:ENSG00000178568|HPRD:02767|Vega:OTTHUMG00000133012 |
| Gene type | protein-coding |
| Map location | 2q33.3-q34 |
| Fetal beta | -0.176 |
| DMG | 1 (# studies) |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 33.331 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04784316 | 2 | 212609384 | ERBB4 | 1.001E-4 | -0.84 | 0.028 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
General gene expression (GTEx)
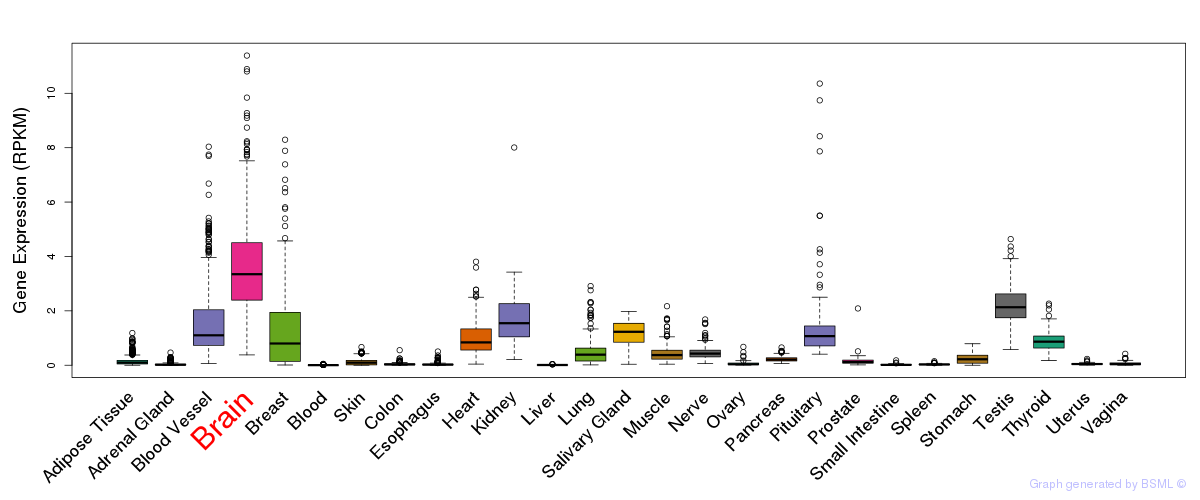
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF2C1 | 0.88 | 0.87 |
| ANKFY1 | 0.88 | 0.90 |
| ZFYVE26 | 0.88 | 0.89 |
| SEC16A | 0.88 | 0.88 |
| TRRAP | 0.87 | 0.87 |
| EP400 | 0.87 | 0.88 |
| ABL1 | 0.87 | 0.85 |
| RNF144A | 0.86 | 0.85 |
| KIAA0355 | 0.86 | 0.88 |
| DIP2A | 0.86 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.59 | -0.70 |
| MT-CO2 | -0.57 | -0.70 |
| C5orf53 | -0.57 | -0.62 |
| HIGD1B | -0.57 | -0.70 |
| IFI27 | -0.57 | -0.67 |
| FXYD1 | -0.56 | -0.66 |
| C1orf54 | -0.54 | -0.75 |
| AF347015.21 | -0.54 | -0.74 |
| AF347015.27 | -0.54 | -0.65 |
| MYL3 | -0.54 | -0.67 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | TAS | neurite (GO term level: 8) | 8383326 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 9419975 |10572067 |11867753 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IDA | 10572067 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007165 | signal transduction | IDA | 10572067 | |
| GO:0008283 | cell proliferation | TAS | 8383326 | |
| GO:0007275 | multicellular organismal development | TAS | 8383326 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8383326 | |
| GO:0016323 | basolateral plasma membrane | IDA | 12646923 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BTC | - | betacellulin | - | HPRD | 11254912 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | - | HPRD,BioGRID | 11825873 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-MS | BioGRID | 16729043 |
| CTGF | CCN2 | HCS24 | IGFBP8 | MGC102839 | NOV2 | connective tissue growth factor | - | HPRD | 12297288 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD,BioGRID | 12175853 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | - | HPRD,BioGRID | 10725395 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | ErbB-4 interacts with CHAPSYN-110. | BIND | 10725395 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | - | HPRD,BioGRID | 10725395 |12175853 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | ErbB-4 interacts with SAP-102. | BIND | 10725395 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 10725395 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | ErbB-4 interacts with PSD-95. | BIND | 10725395 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 10839362 |
| EREG | ER | epiregulin | - | HPRD,BioGRID | 9419975 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-MS | BioGRID | 16729043 |
| HBEGF | DTR | DTS | DTSF | HEGFL | heparin-binding EGF-like growth factor | - | HPRD | 9135143 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | - | HPRD,BioGRID | 11278868 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Affinity Capture-MS | BioGRID | 16729043 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | Reconstituted Complex | BioGRID | 9168115 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | - | HPRD | 7721889 |8062828 |10537356 |
| NRG2 | Don-1 | HRG2 | NTAK | neuregulin 2 | - | HPRD,BioGRID | 9168114 |9168115 |
| NRG3 | HRG3 | pro-NRG3 | neuregulin 3 | - | HPRD | 9275162 |
| NRG4 | DKFZp779N0541 | DKFZp779N1944 | HRG4 | neuregulin 4 | - | HPRD | 10348342 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | Affinity Capture-MS | BioGRID | 16729043 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Affinity Capture-MS | BioGRID | 16729043 |
| RNF41 | MGC45228 | NRDP1 | SBBI03 | ring finger protein 41 | - | HPRD,BioGRID | 11867753 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Affinity Capture-MS | BioGRID | 16729043 |
| SNTB2 | D16S2531E | EST25263 | SNT2B2 | SNT3 | SNTL | syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) | - | HPRD,BioGRID | 10725395 |
| SNTB2 | D16S2531E | EST25263 | SNT2B2 | SNT3 | SNTL | syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) | ErbB-4 interacts with beta 2-syntropin. | BIND | 10725395 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15534001 |16729043 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | - | HPRD,BioGRID | 15023535 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HER2 PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID ERBB NETWORK PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | 22 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | 20 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCERS KINOME BLUE | 21 | 16 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR DN | 91 | 56 | All SZGR 2.0 genes in this pathway |
| LIU TOPBP1 TARGETS | 16 | 8 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 7184 | 7190 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-125/351 | 6482 | 6488 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-130/301 | 1080 | 1086 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-135 | 5566 | 5573 | 1A,m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-137 | 1140 | 1147 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-138 | 1156 | 1162 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-143 | 6806 | 6812 | m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-144 | 7184 | 7190 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-146 | 7692 | 7699 | 1A,m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-19 | 1079 | 1085 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-193 | 91 | 98 | 1A,m8 | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-194 | 7825 | 7831 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-199 | 1034 | 1041 | 1A,m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-205 | 7506 | 7513 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-221/222 | 6627 | 6634 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-23 | 7199 | 7206 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-24* | 6253 | 6260 | 1A,m8 | hsa-miR-189 | GUGCCUACUGAGCUGAUAUCAGU |
| miR-26 | 157 | 163 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-3p | 178 | 184 | m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-323 | 7199 | 7205 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-326 | 5021 | 5028 | 1A,m8 | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-330 | 7266 | 7273 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA | ||||
| miR-362 | 7698 | 7704 | m8 | hsa-miR-362 | AAUCCUUGGAACCUAGGUGUGAGU |
| miR-381 | 7325 | 7331 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-383 | 7782 | 7788 | 1A | hsa-miR-383brain | AGAUCAGAAGGUGAUUGUGGCU |
| miR-495 | 7823 | 7829 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-543 | 7543 | 7549 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCU | ||||
| miR-7 | 73 | 79 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1082 | 1088 | 1A | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.