| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | Two-hybrid | BioGRID | 16189514 |
| AGT | ANHU | FLJ92595 | FLJ97926 | SERPINA8 | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | Two-hybrid | BioGRID | 16189514 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Affinity Capture-MS | BioGRID | 17353931 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Two-hybrid | BioGRID | 16189514 |
| BARD1 | - | BRCA1 associated RING domain 1 | - | HPRD,BioGRID | 12183411 |
| BNIP3L | BNIP3a | NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like | Two-hybrid | BioGRID | 16189514 |
| BTK | AGMX1 | AT | ATK | BPK | IMD1 | MGC126261 | MGC126262 | PSCTK1 | XLA | Bruton agammaglobulinemia tyrosine kinase | - | HPRD,BioGRID | 9201297 |
| C10orf12 | DKFZp564P1916 | FLJ13022 | chromosome 10 open reading frame 12 | Two-hybrid | BioGRID | 16189514 |
| C11orf16 | - | chromosome 11 open reading frame 16 | Two-hybrid | BioGRID | 16189514 |
| C18orf10 | DKFZp586M1523 | HMFN0601 | HsT3006 | L17 | PGs2 | chromosome 18 open reading frame 10 | Two-hybrid | BioGRID | 16189514 |
| C19orf50 | FLJ25480 | MGC2749 | MST096 | MSTP096 | chromosome 19 open reading frame 50 | Two-hybrid | BioGRID | 16189514 |
| C19orf57 | MGC11271 | MGC149720 | chromosome 19 open reading frame 57 | Two-hybrid | BioGRID | 16189514 |
| C1orf71 | FLJ32001 | MGC18089 | chromosome 1 open reading frame 71 | Two-hybrid | BioGRID | 16189514 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | Reconstituted Complex | BioGRID | 9341188 |
| CCDC7 | BioT2-A | BioT2-B | BioT2-C | DKFZp686N0559 | FLJ32762 | RP11-479G22.1 | coiled-coil domain containing 7 | Two-hybrid | BioGRID | 16189514 |
| CCDC91 | DKFZp779L1558 | FLJ11088 | HSD8 | p56 | coiled-coil domain containing 91 | Two-hybrid | BioGRID | 16189514 |
| CD177 | HNA2A | NB1 | PRV1 | CD177 molecule | Two-hybrid | BioGRID | 16189514 |
| CD2BP2 | FWP010 | LIN1 | Snu40 | U5-52K | CD2 (cytoplasmic tail) binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CEACAM5 | CD66e | CEA | DKFZp781M2392 | carcinoembryonic antigen-related cell adhesion molecule 5 | Two-hybrid | BioGRID | 16189514 |
| CETP | HDLCQ10 | cholesteryl ester transfer protein, plasma | Two-hybrid | BioGRID | 16189514 |
| CFDP1 | BCNT | BUCENTAUR | CP27 | SWC5 | Yeti | p97 | craniofacial development protein 1 | Two-hybrid | BioGRID | 16189514 |
| CPSF6 | CFIM | CFIM68 | HPBRII-4 | HPBRII-7 | cleavage and polyadenylation specific factor 6, 68kDa | Two-hybrid | BioGRID | 16189514 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 12459554 |
| CUEDC2 | C10orf66 | MGC2491 | bA18I14.5 | CUE domain containing 2 | Two-hybrid | BioGRID | 16189514 |
| CXADR | CAR | HCAR | coxsackie virus and adenovirus receptor | Two-hybrid | BioGRID | 16189514 |
| DFFA | DFF-45 | DFF1 | ICAD | DNA fragmentation factor, 45kDa, alpha polypeptide | Two-hybrid | BioGRID | 16189514 |
| DMRTB1 | - | DMRT-like family B with proline-rich C-terminal, 1 | Two-hybrid | BioGRID | 16189514 |
| DNAJB3 | HCG3 | MGC26879 | DnaJ (Hsp40) homolog, subfamily B, member 3 | Two-hybrid | BioGRID | 16189514 |
| DYNLL2 | DNCL1B | Dlc2 | MGC17810 | dynein, light chain, LC8-type 2 | Two-hybrid | BioGRID | 16189514 |
| ELAVL3 | DKFZp547J036 | HUC | HUCL | MGC20653 | PLE21 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) | Two-hybrid | BioGRID | 16189514 |
| ELAVL4 | HUD | PNEM | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) | Two-hybrid | BioGRID | 16189514 |
| ELK1 | - | ELK1, member of ETS oncogene family | Reconstituted Complex | BioGRID | 9010223 |
| FAM131C | C1orf117 | FLJ36766 | RP11-5P18.9 | family with sequence similarity 131, member C | Two-hybrid | BioGRID | 16189514 |
| FASN | FAS | MGC14367 | MGC15706 | OA-519 | fatty acid synthase | Two-hybrid | BioGRID | 16189514 |
| FLJ12529 | FLJ39024 | MGC9315 | pre-mRNA cleavage factor I, 59 kDa subunit | Two-hybrid | BioGRID | 16189514 |
| GNPDA1 | GNPDA | GNPI | GPI | HLN | KIAA0060 | glucosamine-6-phosphate deaminase 1 | Two-hybrid | BioGRID | 16189514 |
| GPBP1L1 | RP11-767N6.1 | SP192 | GC-rich promoter binding protein 1-like 1 | Two-hybrid | BioGRID | 16189514 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-MS | BioGRID | 17353931 |
| HERPUD1 | HERP | KIAA0025 | Mif1 | SUP | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | Two-hybrid | BioGRID | 16189514 |
| HMGA1 | HMG-R | HMGA1A | HMGIY | MGC12816 | MGC4242 | MGC4854 | high mobility group AT-hook 1 | Two-hybrid | BioGRID | 16189514 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | Affinity Capture-MS | BioGRID | 17353931 |
| KCNMB1 | K(VCA)beta | SLO-BETA | hslo-beta | potassium large conductance calcium-activated channel, subfamily M, beta member 1 | Two-hybrid | BioGRID | 16189514 |
| KEL | CD238 | ECE3 | Kell blood group, metallo-endopeptidase | Two-hybrid | BioGRID | 16189514 |
| KHDRBS2 | FLJ38664 | MGC26664 | SLM-1 | SLM1 | bA535F17.1 | KH domain containing, RNA binding, signal transduction associated 2 | Two-hybrid | BioGRID | 16189514 |
| KRR1 | HRB2 | RIP-1 | KRR1, small subunit (SSU) processome component, homolog (yeast) | Two-hybrid | BioGRID | 16189514 |
| LILRA3 | CD85E | HM31 | HM43 | ILT6 | LIR-4 | LIR4 | e3 | leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 | Two-hybrid | BioGRID | 16189514 |
| MAGEA11 | MAGE-11 | MAGE11 | MAGEA-11 | MGC10511 | melanoma antigen family A, 11 | Two-hybrid | BioGRID | 16189514 |
| MAPK1IP1L | C14orf32 | MGC23138 | MISS | c14_5346 | mitogen-activated protein kinase 1 interacting protein 1-like | Two-hybrid | BioGRID | 16189514 |
| MATK | CHK | CTK | DKFZp434N1212 | HHYLTK | HYL | HYLTK | Lsk | MGC1708 | MGC2101 | megakaryocyte-associated tyrosine kinase | Two-hybrid | BioGRID | 16189514 |
| MDFI | I-MF | I-mfa | MyoD family inhibitor | Two-hybrid | BioGRID | 16189514 |
| MNS1 | FLJ11222 | FLJ26051 | meiosis-specific nuclear structural 1 | Two-hybrid | BioGRID | 16189514 |
| MRPS18B | C6orf14 | DKFZp564H0223 | HSPC183 | HumanS18a | MRP-S18-2 | MRPS18-2 | PTD017 | S18amt | mitochondrial ribosomal protein S18B | Two-hybrid | BioGRID | 16189514 |
| MSC | ABF-1 | ABF1 | MYOR | bHLHa22 | musculin (activated B-cell factor-1) | Two-hybrid | BioGRID | 16189514 |
| MTCP1 | C6.1B | mature T-cell proliferation 1 | Two-hybrid | BioGRID | 16189514 |
| MTMR9 | C8orf9 | DKFZp434K171 | LIP-STYX | MGC126672 | MTMR8 | myotubularin related protein 9 | Two-hybrid | BioGRID | 16189514 |
| MVK | FLJ96772 | LRBP | MK | MVLK | mevalonate kinase | Two-hybrid | BioGRID | 16189514 |
| MYL6 | ESMLC | LC17-GI | LC17-NM | LC17A | LC17B | MLC1SM | MLC3NM | MLC3SM | myosin, light chain 6, alkali, smooth muscle and non-muscle | Two-hybrid | BioGRID | 16189514 |
| MYO1F | - | myosin IF | Two-hybrid | BioGRID | 16189514 |
| MYOZ2 | C4orf5 | CS-1 | myozenin 2 | Two-hybrid | BioGRID | 16189514 |
| NBPF3 | AE2 | DKFZp434D177 | neuroblastoma breakpoint family, member 3 | Two-hybrid | BioGRID | 16189514 |
| NDUFB1 | CI-SGDH | MNLL | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa | Two-hybrid | BioGRID | 16189514 |
| NDUFV1 | CI-51kD | UQOR1 | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | Two-hybrid | BioGRID | 16189514 |
| NLE1 | FLJ10458 | Nle | notchless homolog 1 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| NPPB | BNP | natriuretic peptide precursor B | Two-hybrid | BioGRID | 16189514 |
| NSUN4 | MGC22960 | RP4-603I14.2 | NOL1/NOP2/Sun domain family, member 4 | Two-hybrid | BioGRID | 16189514 |
| NTNG2 | KIAA0625 | KIAA1857 | LHLL9381 | Lmnt2 | MGC21884 | NTNG1 | bA479K20.1 | netrin G2 | Two-hybrid | BioGRID | 16189514 |
| PDHX | DLDBP | E3BP | OPDX | PDX1 | proX | pyruvate dehydrogenase complex, component X | Two-hybrid | BioGRID | 16189514 |
| PGLS | 6PGL | 6-phosphogluconolactonase | Two-hybrid | BioGRID | 16189514 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Two-hybrid | BioGRID | 16189514 |
| POLR2G | MGC138367 | MGC138369 | RPB7 | hRPB19 | hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G | - | HPRD | 9704926 |
| POU4F1 | BRN3A | FLJ13449 | Oct-T1 | RDC-1 | POU class 4 homeobox 1 | - | HPRD,BioGRID | 12432261 |
| PRTFDC1 | FLJ11888 | HHGP | phosphoribosyl transferase domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| PRUNE2 | A214N16.3 | BMCC1 | BNIPXL | C9orf65 | DKFZp762K117 | FLJ50060 | FLJ54876 | FLJ59118 | KIAA0367 | RP11-58J3.2 | bA214N16.3 | prune homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10322114 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Affinity Capture-MS | BioGRID | 17353931 |
| RAB37 | FLJ30284 | FLJ32507 | RAB37, member RAS oncogene family | Two-hybrid | BioGRID | 16189514 |
| RAD23A | HHR23A | MGC111083 | RAD23 homolog A (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| RALYL | HNRPCL3 | RALY RNA binding protein-like | Two-hybrid | BioGRID | 16189514 |
| RASL11B | MGC2827 | MGC4499 | RAS-like, family 11, member B | Two-hybrid | BioGRID | 16189514 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| RHOXF2 | PEPP-2 | PEPP2 | THG1 | Rhox homeobox family, member 2 | Two-hybrid | BioGRID | 16189514 |
| RMND5B | DKFZp434K0926 | FLJ22318 | required for meiotic nuclear division 5 homolog B (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| RNF183 | FLJ31197 | MGC4734 | ring finger protein 183 | Two-hybrid | BioGRID | 16189514 |
| RP4-691N24.1 | FLJ11792 | KIAA0980 | NLP | dJ691N24.1 | ninein-like | Two-hybrid | BioGRID | 16189514 |
| RPL31 | MGC88191 | ribosomal protein L31 | Two-hybrid | BioGRID | 16189514 |
| RPS15A | FLJ27457 | MGC111208 | S15a | ribosomal protein S15a | Two-hybrid | BioGRID | 16189514 |
| SALL2 | FLJ10414 | FLJ55746 | HSAL2 | KIAA0360 | ZNF795 | p150(Sal2) | sal-like 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| SELI | KIAA1724 | selenoprotein I | Two-hybrid | BioGRID | 16189514 |
| SERP2 | C13orf21 | MGC35505 | RP11-269C23.1 | bA269C23.1 | stress-associated endoplasmic reticulum protein family member 2 | Two-hybrid | BioGRID | 16189514 |
| SF1 | D11S636 | ZFM1 | ZNF162 | splicing factor 1 | - | HPRD,BioGRID | 9660765 |
| SLC1A1 | EAAC1 | EAAT3 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | Two-hybrid | BioGRID | 16189514 |
| SLC22A24 | MGC34821 | solute carrier family 22, member 24 | Two-hybrid | BioGRID | 16189514 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Two-hybrid | BioGRID | 16189514 |
| SMNDC1 | SMNR | SPF30 | survival motor neuron domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SNRPC | FLJ20302 | small nuclear ribonucleoprotein polypeptide C | - | HPRD,BioGRID | 10827180 |
| SSBP2 | DKFZp686F03273 | HSPC116 | single-stranded DNA binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| SUV39H2 | FLJ23414 | KMT1B | suppressor of variegation 3-9 homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | - | HPRD | 9488465 |
| TMSB4Y | MGC26307 | TB4Y | thymosin beta 4, Y-linked | Two-hybrid | BioGRID | 16189514 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | Two-hybrid | BioGRID | 16189514 |
| TRPV5 | CAT2 | ECAC1 | OTRPC3 | transient receptor potential cation channel, subfamily V, member 5 | Two-hybrid | BioGRID | 16189514 |
| TSPAN3 | TM4-A | TM4SF8 | TSPAN-3 | tetraspanin 3 | Two-hybrid | BioGRID | 16189514 |
| TULP2 | TUBL2 | tubby like protein 2 | Two-hybrid | BioGRID | 16189514 |
| VPS72 | CFL1 | Swc2 | TCFL1 | YL-1 | YL1 | vacuolar protein sorting 72 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| WDR37 | FLJ40354 | KIAA0982 | RP11-529L18.2 | WD repeat domain 37 | Two-hybrid | BioGRID | 16189514 |
| WWP1 | AIP5 | DKFZp434D2111 | Tiul1 | hSDRP1 | WW domain containing E3 ubiquitin protein ligase 1 | Two-hybrid | BioGRID | 16189514 |
| WWP2 | AIP2 | WWp2-like | WW domain containing E3 ubiquitin protein ligase 2 | Two-hybrid | BioGRID | 16189514 |
| YY1AP1 | FLJ10875 | FLJ13914 | HCCA1 | HCCA2 | YAP | YY1AP | YY1 associated protein 1 | Two-hybrid | BioGRID | 16189514 |
| ZDHHC3 | DKFZp313D2314 | FLJ20209 | FLJ45940 | GODZ | ZNF373 | zinc finger, DHHC-type containing 3 | Two-hybrid | BioGRID | 16189514 |
| ZNF165 | LD65 | ZSCAN7 | zinc finger protein 165 | Affinity Capture-Western
Two-hybrid | BioGRID | 16189514 |
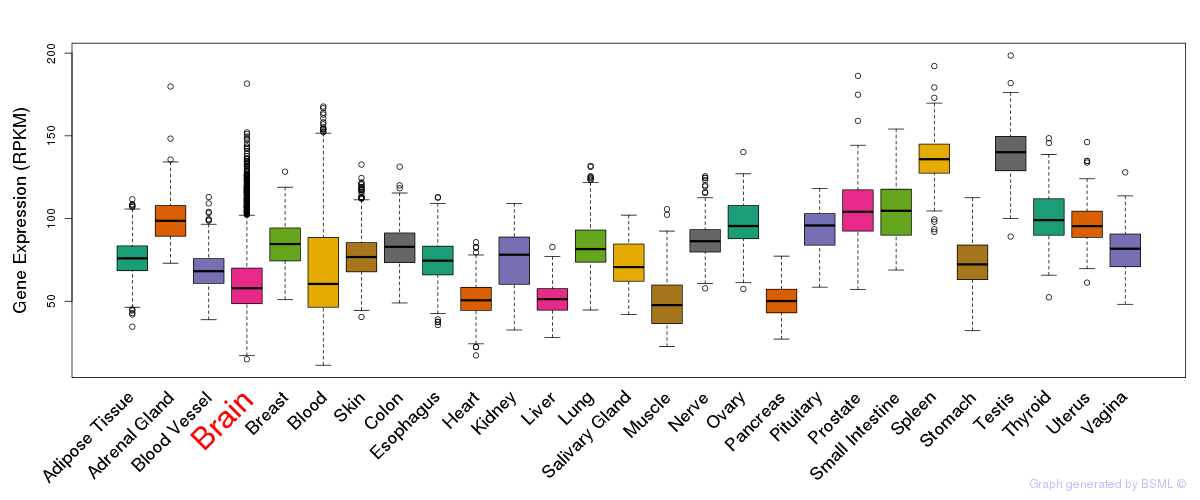
 eQTL annotation
eQTL annotation