Gene Page: ACAP2
Summary ?
| GeneID | 23527 |
| Symbol | ACAP2 |
| Synonyms | CENTB2|CNT-B2 |
| Description | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| Reference | MIM:607766|HGNC:HGNC:16469|Ensembl:ENSG00000114331|HPRD:09681|Vega:OTTHUMG00000155885 |
| Gene type | protein-coding |
| Map location | 3q29 |
| Pascal p-value | 0.077 |
| Fetal beta | 0.754 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cortex Frontal Cortex BA9 Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09474336 | 3 | 195163530 | ACAP2 | 3.1E-8 | -0.013 | 9.43E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6781643 | 3 | 194985678 | ACAP2 | ENSG00000114331.8 | 3.404E-8 | 0 | 178129 | gtex_brain_ba24 |
| rs6791943 | 3 | 194986167 | ACAP2 | ENSG00000114331.8 | 2.912E-7 | 0 | 177640 | gtex_brain_ba24 |
| rs6792029 | 3 | 194986224 | ACAP2 | ENSG00000114331.8 | 2.909E-7 | 0 | 177583 | gtex_brain_ba24 |
| rs6780743 | 3 | 194988441 | ACAP2 | ENSG00000114331.8 | 2.849E-7 | 0 | 175366 | gtex_brain_ba24 |
| rs12695049 | 3 | 194988940 | ACAP2 | ENSG00000114331.8 | 9.02E-8 | 0 | 174867 | gtex_brain_ba24 |
| rs34354069 | 3 | 194989328 | ACAP2 | ENSG00000114331.8 | 1.585E-6 | 0 | 174479 | gtex_brain_ba24 |
| rs7640099 | 3 | 194991220 | ACAP2 | ENSG00000114331.8 | 2.765E-7 | 0 | 172587 | gtex_brain_ba24 |
| rs7640100 | 3 | 194991221 | ACAP2 | ENSG00000114331.8 | 3.911E-7 | 0 | 172586 | gtex_brain_ba24 |
| rs1832667 | 3 | 194992039 | ACAP2 | ENSG00000114331.8 | 7.926E-7 | 0 | 171768 | gtex_brain_ba24 |
| rs7649142 | 3 | 194993295 | ACAP2 | ENSG00000114331.8 | 5.691E-7 | 0 | 170512 | gtex_brain_ba24 |
| rs13072883 | 3 | 194994754 | ACAP2 | ENSG00000114331.8 | 7.612E-7 | 0 | 169053 | gtex_brain_ba24 |
| rs9863901 | 3 | 194996737 | ACAP2 | ENSG00000114331.8 | 5.87E-7 | 0 | 167070 | gtex_brain_ba24 |
| rs200598935 | 3 | 194998809 | ACAP2 | ENSG00000114331.8 | 1.616E-6 | 0 | 164998 | gtex_brain_ba24 |
| rs3087885 | 3 | 194999609 | ACAP2 | ENSG00000114331.8 | 1.724E-6 | 0 | 164198 | gtex_brain_ba24 |
| rs7610022 | 3 | 194999996 | ACAP2 | ENSG00000114331.8 | 5.87E-7 | 0 | 163811 | gtex_brain_ba24 |
| rs2050810 | 3 | 195001730 | ACAP2 | ENSG00000114331.8 | 5.87E-7 | 0 | 162077 | gtex_brain_ba24 |
| rs9823964 | 3 | 195004904 | ACAP2 | ENSG00000114331.8 | 7.612E-7 | 0 | 158903 | gtex_brain_ba24 |
| rs9810077 | 3 | 195005422 | ACAP2 | ENSG00000114331.8 | 7.612E-7 | 0 | 158385 | gtex_brain_ba24 |
| rs3773418 | 3 | 195006791 | ACAP2 | ENSG00000114331.8 | 1.062E-6 | 0 | 157016 | gtex_brain_ba24 |
| rs2410839 | 3 | 195007260 | ACAP2 | ENSG00000114331.8 | 5.87E-7 | 0 | 156547 | gtex_brain_ba24 |
| rs1538767 | 3 | 195007852 | ACAP2 | ENSG00000114331.8 | 1.902E-7 | 0 | 155955 | gtex_brain_ba24 |
| rs9832920 | 3 | 195011869 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 151938 | gtex_brain_ba24 |
| rs9840346 | 3 | 195011895 | ACAP2 | ENSG00000114331.8 | 6.715E-7 | 0 | 151912 | gtex_brain_ba24 |
| rs6778719 | 3 | 195015251 | ACAP2 | ENSG00000114331.8 | 2.028E-7 | 0 | 148556 | gtex_brain_ba24 |
| rs6791601 | 3 | 195019510 | ACAP2 | ENSG00000114331.8 | 1.352E-7 | 0 | 144297 | gtex_brain_ba24 |
| rs6795263 | 3 | 195020188 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 143619 | gtex_brain_ba24 |
| rs13099106 | 3 | 195025263 | ACAP2 | ENSG00000114331.8 | 5.198E-8 | 0 | 138544 | gtex_brain_ba24 |
| rs6437467 | 3 | 195026078 | ACAP2 | ENSG00000114331.8 | 1.025E-6 | 0 | 137729 | gtex_brain_ba24 |
| rs13059843 | 3 | 195028183 | ACAP2 | ENSG00000114331.8 | 3.605E-8 | 0 | 135624 | gtex_brain_ba24 |
| rs56132877 | 3 | 195036583 | ACAP2 | ENSG00000114331.8 | 8.422E-7 | 0 | 127224 | gtex_brain_ba24 |
| rs55927338 | 3 | 195036585 | ACAP2 | ENSG00000114331.8 | 8.422E-7 | 0 | 127222 | gtex_brain_ba24 |
| rs11706264 | 3 | 195037711 | ACAP2 | ENSG00000114331.8 | 8.728E-7 | 0 | 126096 | gtex_brain_ba24 |
| rs13062518 | 3 | 195038287 | ACAP2 | ENSG00000114331.8 | 3.579E-8 | 0 | 125520 | gtex_brain_ba24 |
| rs10933706 | 3 | 195043119 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 120688 | gtex_brain_ba24 |
| rs55672262 | 3 | 195047856 | ACAP2 | ENSG00000114331.8 | 3.587E-8 | 0 | 115951 | gtex_brain_ba24 |
| rs4677825 | 3 | 195047912 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 115895 | gtex_brain_ba24 |
| rs2050809 | 3 | 195048369 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 115438 | gtex_brain_ba24 |
| rs7633265 | 3 | 195048814 | ACAP2 | ENSG00000114331.8 | 3.587E-8 | 0 | 114993 | gtex_brain_ba24 |
| rs13091924 | 3 | 195051136 | ACAP2 | ENSG00000114331.8 | 3.587E-8 | 0 | 112671 | gtex_brain_ba24 |
| rs7638924 | 3 | 195052494 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 111313 | gtex_brain_ba24 |
| rs4677830 | 3 | 195060343 | ACAP2 | ENSG00000114331.8 | 1.024E-6 | 0 | 103464 | gtex_brain_ba24 |
| rs7644002 | 3 | 195064198 | ACAP2 | ENSG00000114331.8 | 1.025E-6 | 0 | 99609 | gtex_brain_ba24 |
| rs13072826 | 3 | 195064755 | ACAP2 | ENSG00000114331.8 | 5.181E-8 | 0 | 99052 | gtex_brain_ba24 |
| rs60767217 | 3 | 195069059 | ACAP2 | ENSG00000114331.8 | 5.184E-8 | 0 | 94748 | gtex_brain_ba24 |
| rs10804414 | 3 | 195072126 | ACAP2 | ENSG00000114331.8 | 5.189E-8 | 0 | 91681 | gtex_brain_ba24 |
| rs11718218 | 3 | 195072425 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 91382 | gtex_brain_ba24 |
| rs7611238 | 3 | 195072918 | ACAP2 | ENSG00000114331.8 | 2.081E-7 | 0 | 90889 | gtex_brain_ba24 |
| rs7634345 | 3 | 195073403 | ACAP2 | ENSG00000114331.8 | 5.19E-8 | 0 | 90404 | gtex_brain_ba24 |
| rs9860389 | 3 | 195074549 | ACAP2 | ENSG00000114331.8 | 1.029E-6 | 0 | 89258 | gtex_brain_ba24 |
| rs7612694 | 3 | 195075672 | ACAP2 | ENSG00000114331.8 | 1.033E-6 | 0 | 88135 | gtex_brain_ba24 |
| rs6437374 | 3 | 195076877 | ACAP2 | ENSG00000114331.8 | 2.199E-12 | 0 | 86930 | gtex_brain_ba24 |
| rs7651757 | 3 | 195077305 | ACAP2 | ENSG00000114331.8 | 3.615E-8 | 0 | 86502 | gtex_brain_ba24 |
| rs60976085 | 3 | 195082175 | ACAP2 | ENSG00000114331.8 | 4.687E-8 | 0 | 81632 | gtex_brain_ba24 |
| rs13324903 | 3 | 195086647 | ACAP2 | ENSG00000114331.8 | 5.172E-8 | 0 | 77160 | gtex_brain_ba24 |
| rs28375276 | 3 | 195088167 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 75640 | gtex_brain_ba24 |
| rs4677834 | 3 | 195093631 | ACAP2 | ENSG00000114331.8 | 3.587E-8 | 0 | 70176 | gtex_brain_ba24 |
| rs58204696 | 3 | 195094042 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 69765 | gtex_brain_ba24 |
| rs72065172 | 3 | 195094137 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 69670 | gtex_brain_ba24 |
| rs9878312 | 3 | 195096404 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 67403 | gtex_brain_ba24 |
| rs4677835 | 3 | 195096995 | ACAP2 | ENSG00000114331.8 | 2.073E-7 | 0 | 66812 | gtex_brain_ba24 |
| rs823302 | 3 | 195104741 | ACAP2 | ENSG00000114331.8 | 3.587E-8 | 0 | 59066 | gtex_brain_ba24 |
| rs823303 | 3 | 195105055 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 58752 | gtex_brain_ba24 |
| rs823304 | 3 | 195106012 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 57795 | gtex_brain_ba24 |
| rs823305 | 3 | 195106167 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 57640 | gtex_brain_ba24 |
| rs823306 | 3 | 195108320 | ACAP2 | ENSG00000114331.8 | 1.191E-7 | 0 | 55487 | gtex_brain_ba24 |
| rs823308 | 3 | 195111916 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 51891 | gtex_brain_ba24 |
| rs823309 | 3 | 195116255 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 47552 | gtex_brain_ba24 |
| rs823297 | 3 | 195120090 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 43717 | gtex_brain_ba24 |
| rs823298 | 3 | 195121072 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 42735 | gtex_brain_ba24 |
| rs823299 | 3 | 195122167 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 41640 | gtex_brain_ba24 |
| rs823300 | 3 | 195122328 | ACAP2 | ENSG00000114331.8 | 3.594E-8 | 0 | 41479 | gtex_brain_ba24 |
| rs823301 | 3 | 195123495 | ACAP2 | ENSG00000114331.8 | 3.594E-8 | 0 | 40312 | gtex_brain_ba24 |
| rs823310 | 3 | 195127093 | ACAP2 | ENSG00000114331.8 | 4.64E-8 | 0 | 36714 | gtex_brain_ba24 |
| rs1094677 | 3 | 195129744 | ACAP2 | ENSG00000114331.8 | 3.594E-8 | 0 | 34063 | gtex_brain_ba24 |
| rs823314 | 3 | 195132402 | ACAP2 | ENSG00000114331.8 | 3.556E-8 | 0 | 31405 | gtex_brain_ba24 |
| rs147522534 | 3 | 195133475 | ACAP2 | ENSG00000114331.8 | 7.199E-8 | 0 | 30332 | gtex_brain_ba24 |
| rs823313 | 3 | 195136251 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 27556 | gtex_brain_ba24 |
| rs823312 | 3 | 195139118 | ACAP2 | ENSG00000114331.8 | 1.023E-6 | 0 | 24689 | gtex_brain_ba24 |
| rs823311 | 3 | 195141029 | ACAP2 | ENSG00000114331.8 | 3.415E-8 | 0 | 22778 | gtex_brain_ba24 |
| rs2791603 | 3 | 195142939 | ACAP2 | ENSG00000114331.8 | 3.125E-8 | 0 | 20868 | gtex_brain_ba24 |
| rs4677671 | 3 | 195148149 | ACAP2 | ENSG00000114331.8 | 1.197E-6 | 0 | 15658 | gtex_brain_ba24 |
| rs9831944 | 3 | 195149742 | ACAP2 | ENSG00000114331.8 | 1.221E-6 | 0 | 14065 | gtex_brain_ba24 |
| rs73080837 | 3 | 195152149 | ACAP2 | ENSG00000114331.8 | 1.254E-6 | 0 | 11658 | gtex_brain_ba24 |
| rs73080839 | 3 | 195152167 | ACAP2 | ENSG00000114331.8 | 1.254E-6 | 0 | 11640 | gtex_brain_ba24 |
| rs57316916 | 3 | 195156861 | ACAP2 | ENSG00000114331.8 | 1.46E-6 | 0 | 6946 | gtex_brain_ba24 |
| rs200616554 | 3 | 195158757 | ACAP2 | ENSG00000114331.8 | 6.298E-7 | 0 | 5050 | gtex_brain_ba24 |
| rs201243710 | 3 | 195158759 | ACAP2 | ENSG00000114331.8 | 1.121E-6 | 0 | 5048 | gtex_brain_ba24 |
| rs1105165 | 3 | 195163917 | ACAP2 | ENSG00000114331.8 | 6.992E-8 | 0 | -110 | gtex_brain_ba24 |
| rs9288747 | 3 | 195165420 | ACAP2 | ENSG00000114331.8 | 9.832E-7 | 0 | -1613 | gtex_brain_ba24 |
| rs6437374 | 3 | 195076877 | ACAP2 | ENSG00000114331.8 | 3.484E-8 | 0 | 86930 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
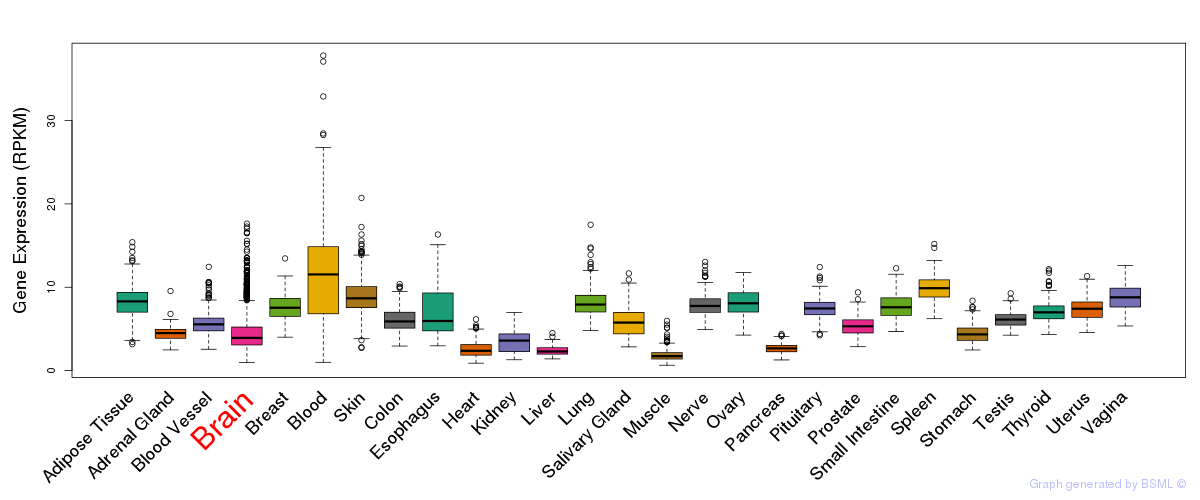
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EHMT1 | 0.98 | 0.95 |
| PHF2 | 0.98 | 0.97 |
| ZBED4 | 0.98 | 0.97 |
| SMARCA4 | 0.97 | 0.96 |
| PHF12 | 0.97 | 0.97 |
| POGZ | 0.97 | 0.97 |
| ZXDC | 0.97 | 0.97 |
| ZNF646 | 0.97 | 0.96 |
| ILF3 | 0.97 | 0.97 |
| ARID1A | 0.96 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.72 | -0.79 |
| AF347015.31 | -0.71 | -0.91 |
| HLA-F | -0.70 | -0.73 |
| AF347015.27 | -0.70 | -0.88 |
| S100B | -0.70 | -0.84 |
| AIFM3 | -0.70 | -0.74 |
| MT-CO2 | -0.69 | -0.90 |
| ALDOC | -0.69 | -0.69 |
| B2M | -0.68 | -0.79 |
| FXYD1 | -0.68 | -0.86 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL DN | 42 | 25 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING SCAR UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |