Gene Page: PADI4
Summary ?
| GeneID | 23569 |
| Symbol | PADI4 |
| Synonyms | PAD|PAD4|PADI5|PDI4|PDI5 |
| Description | peptidyl arginine deiminase, type IV |
| Reference | MIM:605347|HGNC:HGNC:18368|Ensembl:ENSG00000159339|HPRD:05635|Vega:OTTHUMG00000002371 |
| Gene type | protein-coding |
| Map location | 1p36.13 |
| Pascal p-value | 0.513 |
| Fetal beta | 0.16 |
| eGene | Anterior cingulate cortex BA24 Cortex Frontal Cortex BA9 Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11203367 | chr1 | 17657615 | PADI4 | 23569 | 0.02 | cis | ||
| rs1635597 | chr1 | 17659555 | PADI4 | 23569 | 0.03 | cis | ||
| rs1748035 | chr1 | 17661995 | PADI4 | 23569 | 0.03 | cis | ||
| rs11203368 | chr1 | 17666507 | PADI4 | 23569 | 0.05 | cis | ||
| rs12156052 | chr8 | 14903733 | PADI4 | 23569 | 0.11 | trans | ||
| rs114195833 | 1 | 17615542 | PADI4 | ENSG00000159339.9 | 2.041E-6 | 0 | -19148 | gtex_brain_ba24 |
| rs78138254 | 1 | 17631911 | PADI4 | ENSG00000159339.9 | 4.751E-7 | 0 | -2779 | gtex_brain_ba24 |
| rs34470277 | 1 | 17643054 | PADI4 | ENSG00000159339.9 | 1.12E-7 | 0 | 8364 | gtex_brain_ba24 |
| rs35412937 | 1 | 17644463 | PADI4 | ENSG00000159339.9 | 1.007E-7 | 0 | 9773 | gtex_brain_ba24 |
| rs13375202 | 1 | 17651637 | PADI4 | ENSG00000159339.9 | 2.262E-7 | 0 | 16947 | gtex_brain_ba24 |
| rs4920595 | 1 | 17652294 | PADI4 | ENSG00000159339.9 | 2.233E-7 | 0 | 17604 | gtex_brain_ba24 |
| rs12035527 | 1 | 17658337 | PADI4 | ENSG00000159339.9 | 1.533E-6 | 0 | 23647 | gtex_brain_ba24 |
| rs4506507 | 1 | 17659176 | PADI4 | ENSG00000159339.9 | 2.121E-6 | 0 | 24486 | gtex_brain_ba24 |
| rs12737739 | 1 | 17660890 | PADI4 | ENSG00000159339.9 | 2.219E-7 | 0 | 26200 | gtex_brain_ba24 |
| rs11587827 | 1 | 17671382 | PADI4 | ENSG00000159339.9 | 1.52E-6 | 0 | 36692 | gtex_brain_ba24 |
| rs2301888 | 1 | 17672730 | PADI4 | ENSG00000159339.9 | 4.754E-8 | 0 | 38040 | gtex_brain_ba24 |
| rs2240339 | 1 | 17674108 | PADI4 | ENSG00000159339.9 | 8.063E-8 | 0 | 39418 | gtex_brain_ba24 |
| rs2240338 | 1 | 17674185 | PADI4 | ENSG00000159339.9 | 8.063E-8 | 0 | 39495 | gtex_brain_ba24 |
| rs2240336 | 1 | 17674402 | PADI4 | ENSG00000159339.9 | 9.905E-7 | 0 | 39712 | gtex_brain_ba24 |
| rs2240335 | 1 | 17674537 | PADI4 | ENSG00000159339.9 | 4.628E-8 | 0 | 39847 | gtex_brain_ba24 |
Section II. Transcriptome annotation
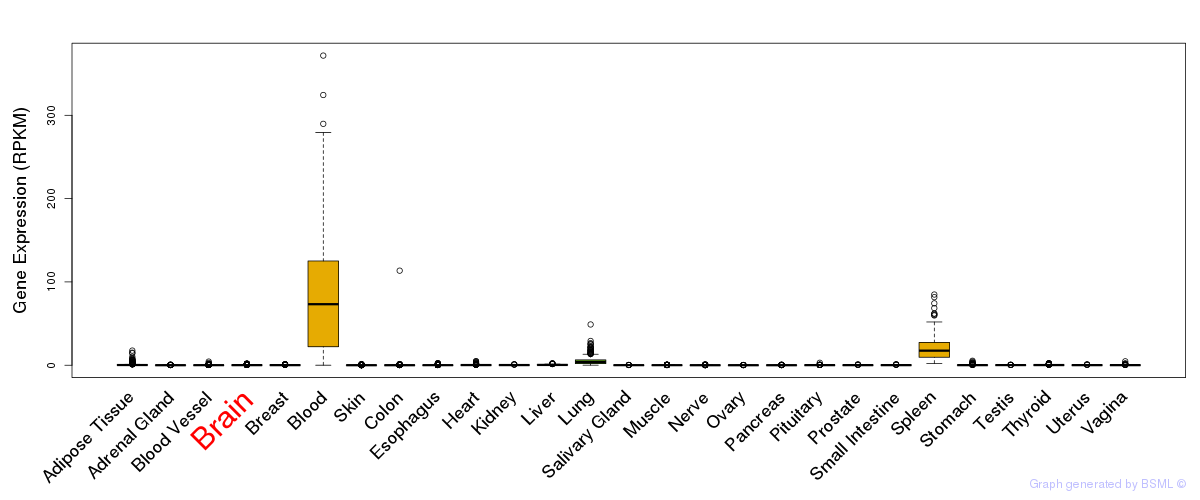
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| PEPPER CHRONIC LYMPHOCYTIC LEUKEMIA UP | 33 | 28 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 DN | 78 | 44 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |