Gene Page: HPRT1
Summary ?
| GeneID | 3251 |
| Symbol | HPRT1 |
| Synonyms | HGPRT|HPRT |
| Description | hypoxanthine phosphoribosyltransferase 1 |
| Reference | MIM:308000|HGNC:HGNC:5157|Ensembl:ENSG00000165704|HPRD:02388|Vega:OTTHUMG00000022452 |
| Gene type | protein-coding |
| Map location | Xq26.1 |
| Sherlock p-value | 0.133 |
| Fetal beta | -2.205 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0044 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02804166 | X | 133506996 | HPRT1 | 0.014 | -4.136 | DMG:vanEijk_2014 | |
| cg10584819 | X | 133593670 | HPRT1 | 0.004 | -4.563 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-1983537 | 0 | HPRT1 | 3251 | 0.17 | trans |
Section II. Transcriptome annotation
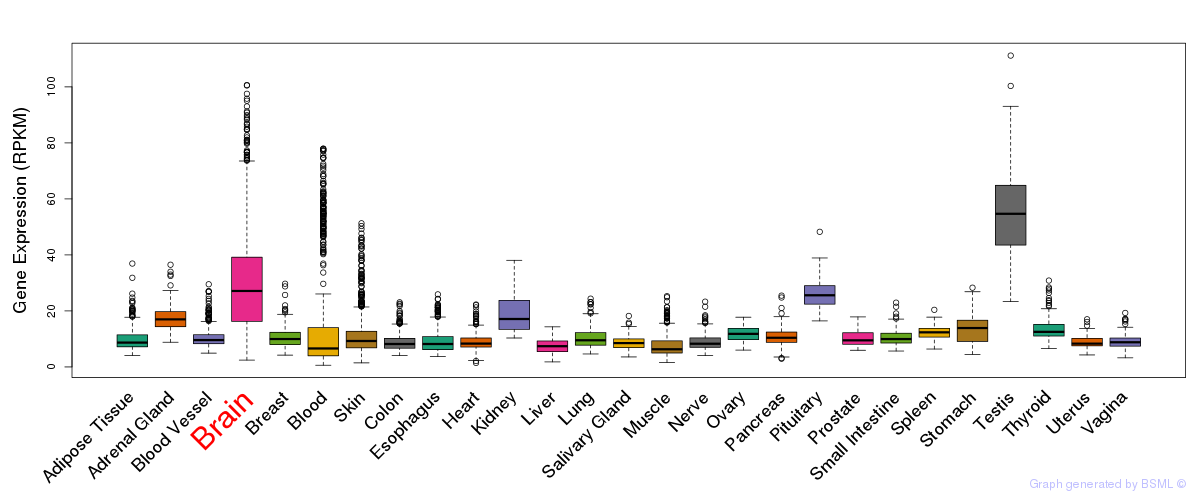
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IDA | 10360366 | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0004422 | hypoxanthine phosphoribosyltransferase activity | EXP | 6300847 | |
| GO:0004422 | hypoxanthine phosphoribosyltransferase activity | IDA | 6300847 |9521733 | |
| GO:0016757 | transferase activity, transferring glycosyl groups | IEA | - | |
| GO:0042803 | protein homodimerization activity | IPI | 8044844 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045964 | positive regulation of dopamine metabolic process | IMP | dopamine (GO term level: 10) | 8643611 |
| GO:0009116 | nucleoside metabolic process | IEA | - | |
| GO:0006166 | purine ribonucleoside salvage | IMP | 9824441 | |
| GO:0006164 | purine nucleotide biosynthetic process | IMP | 9824441 | |
| GO:0007610 | behavior | IMP | 16138897 | |
| GO:0051289 | protein homotetramerization | IDA | 9521733 | |
| GO:0046100 | hypoxanthine metabolic process | IMP | 9824441 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 6300847 | |
| GO:0005737 | cytoplasm | IDA | 6300847 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG DRUG METABOLISM OTHER ENZYMES | 51 | 27 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NUCLEOTIDES | 72 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE SALVAGE | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE METABOLISM | 33 | 24 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| AIYAR COBRA1 TARGETS DN | 29 | 18 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER UP | 66 | 42 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL | 28 | 22 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| FERRANDO HOX11 NEIGHBORS | 23 | 13 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS UP | 112 | 65 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-410 | 345 | 351 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-494 | 484 | 490 | 1A | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.