Gene Page: APP
Summary ?
| GeneID | 351 |
| Symbol | APP |
| Synonyms | AAA|ABETA|ABPP|AD1|APPI|CTFgamma|CVAP|PN-II|PN2 |
| Description | amyloid beta precursor protein |
| Reference | MIM:104760|HGNC:HGNC:620|Ensembl:ENSG00000142192|HPRD:00100|Vega:OTTHUMG00000078438 |
| Gene type | protein-coding |
| Map location | 21q21.3 |
| Pascal p-value | 0.079 |
| Sherlock p-value | 0.726 |
| Fetal beta | -1.23 |
| DMG | 1 (# studies) |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 8.0457 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13169373 | 21 | 27542843 | APP | -0.027 | 0.32 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
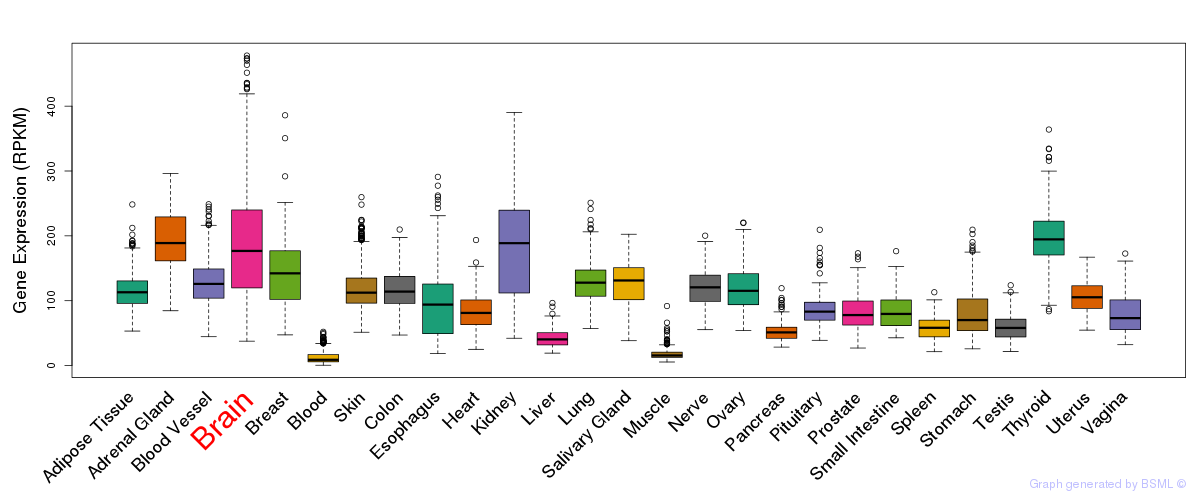
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004867 | serine-type endopeptidase inhibitor activity | IDA | 10652580 | |
| GO:0004867 | serine-type endopeptidase inhibitor activity | NAS | 11279603 | |
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0005507 | copper ion binding | IEA | - | |
| GO:0008201 | heparin binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0033130 | acetylcholine receptor binding | IPI | 10681545 | |
| GO:0042802 | identical protein binding | IPI | 16286452 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007219 | Notch signaling pathway | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0006878 | cellular copper ion homeostasis | TAS | 15910549 | |
| GO:0006897 | endocytosis | IEA | - | |
| GO:0050905 | neuromuscular process | NAS | 7593229 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 18029348 | |
| GO:0005576 | extracellular region | EXP | 2110384 | |
| GO:0005576 | extracellular region | TAS | 10806211 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0009986 | cell surface | IDA | 7593229 | |
| GO:0005905 | coated pit | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0005887 | integral to plasma membrane | TAS | 10806211 | |
| GO:0031093 | platelet alpha granule lumen | EXP | 2110384 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD | 11279131 |
| ACHE | ARACHE | N-ACHE | YT | acetylcholinesterase (Yt blood group) | - | HPRD,BioGRID | 12427014 |
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | - | HPRD,BioGRID | 8887653 |
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | APPswe interacts with X11-alpha. | BIND | 12849748 |14756819 |
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | Beta-APP interacts with X11. | BIND | 8887653 |
| APBA2 | D15S1518E | HsT16821 | LIN-10 | MGC99508 | MGC:14091 | MINT2 | X11L | amyloid beta (A4) precursor protein-binding, family A, member 2 | - | HPRD,BioGRID | 9890987 |
| APBA3 | MGC:15815 | X11L2 | mint3 | amyloid beta (A4) precursor protein-binding, family A, member 3 | - | HPRD,BioGRID | 10049767 |
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8855266 |8887653 |9045663 |9837937 |10081969 |
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | - | HPRD | 8887653 |8955346 |9045663 |9799084 |9837937 |11085987 |
| APBB2 | DKFZp434E033 | FE65L | FE65L1 | MGC35575 | amyloid beta (A4) precursor protein-binding, family B, member 2 | - | HPRD,BioGRID | 8855266 |14527950 |
| APBB3 | FE65L2 | MGC150555 | MGC87674 | SRA | amyloid beta (A4) precursor protein-binding, family B, member 3 | - | HPRD,BioGRID | 10081969|14527950 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD | 10559559 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | APP (Abeta) isoform b forms homodimers, homotrimers and oligomers. | BIND | 15834427 |
| APPBP2 | HS.84084 | KIAA0228 | PAT1 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | - | HPRD,BioGRID | 9843960 |
| BCAP31 | 6C6-AG | BAP31 | CDM | DXS1357E | B-cell receptor-associated protein 31 | - | HPRD,BioGRID | 12529377 |
| BGN | DSPG1 | PG-S1 | PGI | SLRR1A | biglycan | - | HPRD,BioGRID | 7793988 |
| BLMH | BH | BMH | bleomycin hydrolase | - | HPRD,BioGRID | 10973933 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | - | HPRD,BioGRID | 11378243 |
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | - | HPRD | 15123740 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD | 10911620 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 9553108 |
| CLSTN1 | ALC-ALPHA | CSTN1 | FLJ32258 | KIAA0911 | PIK3CD | XB31alpha | alcalpha1 | alcalpha2 | calsyntenin 1 | - | HPRD,BioGRID | 15037614 |
| COL1A2 | OI4 | collagen, type I, alpha 2 | - | HPRD | 7688497 |
| COL25A1 | CLAC | CLACP | collagen, type XXV, alpha 1 | - | HPRD,BioGRID | 11927537 |
| COL4A1 | arresten | collagen, type IV, alpha 1 | - | HPRD | 9136074 |
| COL4A2 | DKFZp686I14213 | FLJ22259 | collagen, type IV, alpha 2 | - | HPRD | 9136074 |
| COL4A3 | - | collagen, type IV, alpha 3 (Goodpasture antigen) | - | HPRD | 9136074 |
| COL4A5 | ASLN | ATS | CA54 | MGC167109 | MGC42377 | collagen, type IV, alpha 5 | - | HPRD | 9136074 |
| COL4A6 | MGC88184 | collagen, type IV, alpha 6 | - | HPRD | 9136074 |
| DAB1 | - | disabled homolog 1 (Drosophila) | - | HPRD,BioGRID | 10460257 |
| DAB1 | - | disabled homolog 1 (Drosophila) | - | HPRD | 9837937 |10460257 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | Reconstituted Complex | BioGRID | 11247302 |
| F12 | HAE3 | HAEX | HAF | coagulation factor XII (Hageman factor) | - | HPRD | 11589915 |
| FBLN1 | FBLN | fibulin 1 | - | HPRD,BioGRID | 11238726 |
| GAPDH | G3PD | GAPD | MGC88685 | glyceraldehyde-3-phosphate dehydrogenase | - | HPRD | 8473906 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 12485888 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | - | HPRD,BioGRID | 10329371 |
| HADHB | ECHB | MGC87480 | MSTP029 | TP-BETA | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | - | HPRD | 11430884 |
| HMOX2 | HO-2 | heme oxygenase (decycling) 2 | - | HPRD | 12057765 |
| HSD17B10 | 17b-HSD10 | ABAD | CAMR | ERAB | HADH2 | HCD2 | MHBD | MRX17 | MRX31 | MRXS10 | SCHAD | hydroxysteroid (17-beta) dehydrogenase 10 | - | HPRD,BioGRID | 9338779 |10371197 |
| HSPG2 | PLC | PRCAN | SJA | SJS | SJS1 | heparan sulfate proteoglycan 2 | - | HPRD | 9136074 |
| HTRA2 | OMI | PARK13 | PRSS25 | HtrA serine peptidase 2 | HtrA2 interacts with APP. | BIND | 15036614 |
| IDE | FLJ35968 | INSULYSIN | insulin-degrading enzyme | - | HPRD | 9830016 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | - | HPRD | 12888553 |
| KLC1 | KLC | KNS2 | KNS2A | MGC15245 | kinesin light chain 1 | - | HPRD,BioGRID | 11144355 |
| KNG1 | BDK | KNG | kininogen 1 | - | HPRD | 12090937 |
| LAMA1 | LAMA | laminin, alpha 1 | - | HPRD | 9136074 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD | 7543026 |12212791 |
| LRP1B | LRP-DIT | LRPDIT | low density lipoprotein-related protein 1B (deleted in tumors) | - | HPRD | 15126508 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | JIP1 interacts with APP. | BIND | 11724784 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | - | HPRD | 11517249 |11724784 |
| NAE1 | A-116A10.1 | APPBP1 | HPP1 | ula-1 | NEDD8 activating enzyme E1 subunit 1 | The carboxy terminus of Amyloid Precursor Protein (APP) mediates an interaction with APP-BP1. | BIND | 8626687 |
| NAE1 | A-116A10.1 | APPBP1 | HPP1 | ula-1 | NEDD8 activating enzyme E1 subunit 1 | - | HPRD,BioGRID | 8626687 |
| NCSTN | APH2 | KIAA0253 | nicastrin | - | HPRD,BioGRID | 10993067 |
| NID1 | NID | nidogen 1 | - | HPRD | 11376898 |
| NUMB | S171 | numb homolog (Drosophila) | - | HPRD | 12011466 |
| PRSS2 | MGC111183 | MGC120174 | TRY2 | TRY8 | TRYP2 | protease, serine, 2 (trypsin 2) | - | HPRD | 8478942 |
| PRSS3 | MTG | PRSS4 | TRY3 | TRY4 | protease, serine, 3 | - | HPRD | 11827488 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | - | HPRD | 10593990 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | APPswe interacts with PS-1delta-9. | BIND | 14756819 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | PS1 interacts with APP via the APP-C100 fragment. | BIND | 10801777 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | PS1 interacts with APP751. | BIND | 9223340 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | PS1 interacts with APP695. | BIND | 9223340 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | PS1 interacts with APP. | BIND | 15448688 |
| PSEN2 | AD3L | AD4 | PS2 | STM2 | presenilin 2 (Alzheimer disease 4) | PS2 interacts with APP. | BIND | 9223340 |
| SERPINA3 | AACT | ACT | GIG24 | GIG25 | MGC88254 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | - | HPRD | 2190106|10048303 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 9045663 |11877420 |
| SHC3 | N-Shc | NSHC | RAI | SHCC | SHC (Src homology 2 domain containing) transforming protein 3 | - | HPRD | 11877420 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD,BioGRID | 9163350 |
| SNCB | - | synuclein, beta | - | HPRD,BioGRID | 9163350 |
| SPON1 | KIAA0762 | MGC10724 | f-spondin | spondin 1, extracellular matrix protein | - | HPRD | 14983046 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | TGF-beta-1 interacts with Amyloid-beta. This interaction was modelled on a demonstrated interaction between TGF-beta-1 from an unspecified species and consensus Amyloid-beta peptides. | BIND | 12867422 |
| TGFB2 | MGC116892 | TGF-beta2 | transforming growth factor, beta 2 | TGF-beta-2 interacts with Amyloid-beta. This interaction was modelled on a demonstrated interaction between TGF-beta-2 from an unspecified species and consensus Amyloid-beta peptides. | BIND | 12867422 |
| TM2D1 | BBP | TM2 domain containing 1 | - | HPRD | 11278849 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | - | HPRD | 11278849 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P35ALZHEIMERS PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PLATELETAPP PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| PID GLYPICAN 1PATHWAY | 27 | 20 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | 23 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED NFKB ACTIVATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME THE NLRP3 INFLAMMASOME | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLAMMASOMES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN | 47 | 32 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS DN | 66 | 39 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8 | 86 | 57 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| CHENG RESPONSE TO NICKEL ACETATE | 45 | 29 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER DN | 74 | 34 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN | 50 | 35 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH FLT3 ITD | 40 | 22 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP | 101 | 66 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
| WARTERS IR RESPONSE 5GY | 47 | 23 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 243 | 250 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| hsa-miR-101 | UACAGUACUGUGAUAACUGAAG | ||||
| miR-144 | 532 | 538 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG | ||||
| miR-15/16/195/424/497 | 544 | 550 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-153 | 458 | 464 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 710 | 716 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-185 | 288 | 294 | m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-324-5p | 393 | 399 | m8 | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-381 | 325 | 331 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-383 | 451 | 457 | 1A | hsa-miR-383brain | AGAUCAGAAGGUGAUUGUGGCU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 709 | 715 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.