Gene Page: ITGB3
Summary ?
| GeneID | 3690 |
| Symbol | ITGB3 |
| Synonyms | BDPLT16|BDPLT2|CD61|GP3A|GPIIIa|GT |
| Description | integrin subunit beta 3 |
| Reference | MIM:173470|HGNC:HGNC:6156|Ensembl:ENSG00000259207|HPRD:01428|Vega:OTTHUMG00000171956 |
| Gene type | protein-coding |
| Map location | 17q21.32 |
| Pascal p-value | 0.608 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1411 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
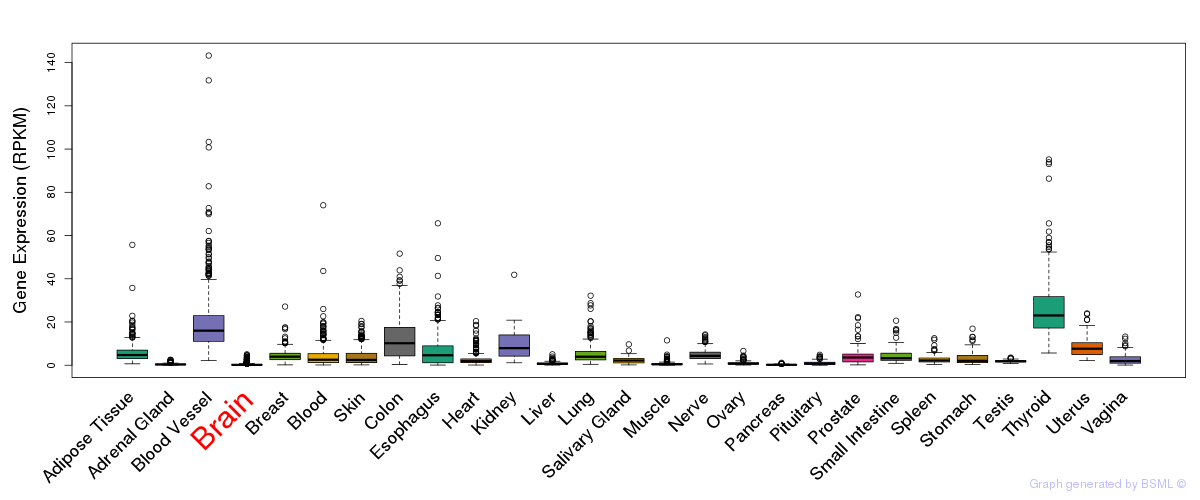
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005178 | integrin binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 11606749 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0007155 | cell adhesion | TAS | 2452834 |10429193 | |
| GO:0007229 | integrin-mediated signaling pathway | IEA | - | |
| GO:0007596 | blood coagulation | TAS | 1438206 | |
| GO:0007044 | cell-substrate junction assembly | IEA | - | |
| GO:0030334 | regulation of cell migration | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009897 | external side of plasma membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 3607284 |10605720 | |
| GO:0008305 | integrin complex | TAS | 1438206 | |
| GO:0031092 | platelet alpha granule membrane | EXP | 3607284 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANGPTL3 | ANGPT5 | angiopoietin-like 3 | - | HPRD | 11877390 |
| CD36 | CHDS7 | FAT | GP3B | GP4 | GPIV | PASIV | SCARB3 | CD36 molecule (thrombospondin receptor) | Affinity Capture-Western | BioGRID | 11238109 |
| CIB1 | CIB | KIP | KIP1 | SIP2-28 | calcium and integrin binding 1 (calmyrin) | - | HPRD,BioGRID | 9030514 |
| FGA | Fib2 | MGC119422 | MGC119423 | MGC119425 | fibrinogen alpha chain | - | HPRD | 11460491 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | Two-hybrid | BioGRID | 11807098 |
| FLNB | ABP-278 | AOI | DKFZp686A1668 | DKFZp686O033 | FH1 | FLN1L | LRS1 | SCT | TABP | TAP | filamin B, beta (actin binding protein 278) | Two-hybrid | BioGRID | 11807098 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | - | HPRD,BioGRID | 12372433 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | Integrin-beta-3 interacts with ILK. | BIND | 12372433 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | Integrin-alphaIIb interacts with Integrin-beta3. | BIND | 8132607 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | - | HPRD | 1429573 |
| ITGAV | CD51 | DKFZp686A08142 | MSK8 | VNRA | integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) | Integrin-alphaV interacts with Integrin-beta3. | BIND | 1693624 |2138612 |
| ITGAV | CD51 | DKFZp686A08142 | MSK8 | VNRA | integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) | - | HPRD,BioGRID | 11884718 |
| ITGB3BP | CENP-R | CENPR | HSU37139 | NRIF3 | TAP20 | integrin beta 3 binding protein (beta3-endonexin) | - | HPRD,BioGRID | 7593198 |11864709 |
| MYO10 | FLJ10639 | FLJ21066 | FLJ22268 | FLJ43256 | KIAA0799 | MGC131988 | myosin X | Integrin-beta-3 interacts with Myo10. This interaction was modeled on a demonstrated interaction between monkey integrin-beta-3 and bovine Myo10. | BIND | 15156152 |
| NID1 | NID | nidogen 1 | - | HPRD | 8831898 |
| P2RY2 | HP2U | MGC20088 | MGC40010 | P2RU1 | P2U | P2U1 | P2UR | P2Y2 | P2Y2R | purinergic receptor P2Y, G-protein coupled, 2 | - | HPRD,BioGRID | 11331301 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | Affinity Capture-Western | BioGRID | 11953315 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Affinity Capture-Western | BioGRID | 10964931 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | Co-localization | BioGRID | 10982404 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | FAK interacts with integrin beta 3. | BIND | 11927607 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9169439 |11927607 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 8649427 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 11683411 |
| PXN | FLJ16691 | paxillin | Reconstituted Complex | BioGRID | 11683411 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Integrin-beta-3 interacts with Shc. | BIND | 10964917 |
| TENC1 | C1-TEN | C1TEN | DKFZp686D13244 | FLJ16320 | KIAA1075 | TNS2 | tensin like C1 domain containing phosphatase (tensin 2) | - | HPRD | 12606711 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | Reconstituted Complex | BioGRID | 2478219 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | - | HPRD,BioGRID | 10497223 |11932255 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EDG1 PATHWAY | 27 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN CS PATHWAY | 26 | 16 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN3 PATHWAY | 43 | 25 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID NECTIN PATHWAY | 30 | 20 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P1 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME PECAM1 INTERACTIONS | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL TRANSDUCTION BY L1 | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP DN | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| DORSEY GAB2 TARGETS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C5 | 21 | 11 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 DN | 78 | 44 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES | 148 | 83 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 1063 | 1069 | m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-1/206 | 407 | 413 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-125/351 | 699 | 705 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-30-5p | 65 | 72 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-495 | 1945 | 1951 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.