Gene Page: LRP1
Summary ?
| GeneID | 4035 |
| Symbol | LRP1 |
| Synonyms | A2MR|APOER|APR|CD91|IGFBP3R|LRP|LRP1A|TGFBR5 |
| Description | LDL receptor related protein 1 |
| Reference | MIM:107770|HGNC:HGNC:6692|Ensembl:ENSG00000123384|HPRD:00138|Vega:OTTHUMG00000044412 |
| Gene type | protein-coding |
| Map location | 12q13.3 |
| Pascal p-value | 6.808E-4 |
| Sherlock p-value | 0.991 |
| TADA p-value | 0.008 |
| Fetal beta | 1.604 |
| Support | CELL METABOLISM G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Girard_2011 | Whole Exome Sequencing analysis | The data set included 15 DNMs found from the exomes of 14 schizophrenia probands and their parents. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.7051 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LRP1 | chr12 | 57579450 | C | A | NM_002332 | p.2200Y>* | nonsense | Schizophrenia | DNM:Girard_2011 |
Section II. Transcriptome annotation
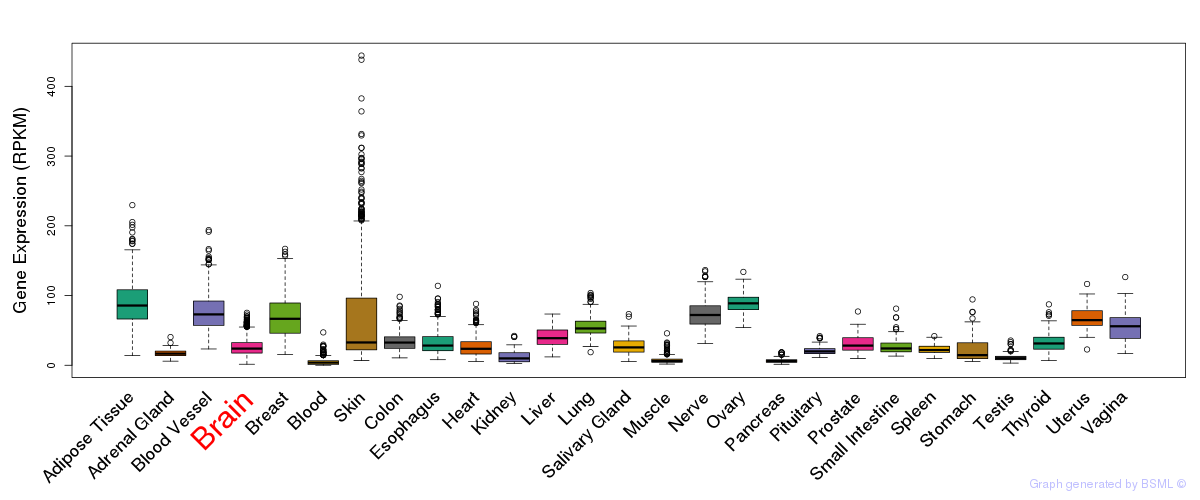
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COL3A1 | 0.90 | 0.64 |
| OLFML3 | 0.87 | 0.73 |
| C7 | 0.86 | 0.73 |
| BMP5 | 0.86 | 0.59 |
| ASPN | 0.85 | 0.74 |
| OGN | 0.82 | 0.78 |
| PCOLCE | 0.81 | 0.48 |
| AL137026.3 | 0.81 | 0.54 |
| SERPIND1 | 0.80 | 0.59 |
| COL1A2 | 0.80 | 0.67 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.33 | -0.25 | -0.19 |
| ACADVL | -0.24 | -0.27 |
| AF347015.26 | -0.24 | -0.19 |
| S100A1 | -0.24 | -0.12 |
| AF347015.8 | -0.24 | -0.19 |
| MT-CO2 | -0.24 | -0.18 |
| AF347015.2 | -0.24 | -0.20 |
| AF347015.27 | -0.24 | -0.17 |
| AF347015.15 | -0.23 | -0.17 |
| AC018755.7 | -0.23 | -0.15 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 10880251 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005509 | calcium ion binding | TAS | 3266596 | |
| GO:0005319 | lipid transporter activity | TAS | 3266596 | |
| GO:0008034 | lipoprotein binding | TAS | 2779654 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008283 | cell proliferation | TAS | 3266596 | |
| GO:0006629 | lipid metabolic process | TAS | 3266596 | |
| GO:0006897 | endocytosis | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030178 | negative regulation of Wnt receptor signaling pathway | IEA | - | |
| GO:0043277 | apoptotic cell clearance | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | TAS | 2779654 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005905 | coated pit | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 3266596 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| A2M | CPAMD5 | DKFZp779B086 | FWP007 | S863-7 | alpha-2-macroglobulin | - | HPRD,BioGRID | 10652313 |
| ANAPC10 | APC10 | DKFZp564L0562 | DOC1 | anaphase promoting complex subunit 10 | - | HPRD,BioGRID | 10827173 |
| APBA2 | D15S1518E | HsT16821 | LIN-10 | MGC99508 | MGC:14091 | MINT2 | X11L | amyloid beta (A4) precursor protein-binding, family A, member 2 | Two-hybrid | BioGRID | 10827173 |
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | - | HPRD,BioGRID | 9837937 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD | 11421580 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 7543026 |12212791 |
| C3 | ARMD9 | ASP | CPAMD1 | complement component 3 | - | HPRD | 10608878 |
| C4BPA | C4BP | PRP | complement component 4 binding protein, alpha | - | HPRD | 11705989 |
| CALCR | CRT | CTR | CTR1 | calcitonin receptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 12821648 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | - | HPRD,BioGRID | 12821648 |14980518 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | - | HPRD,BioGRID | 14980518 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD | 9804823 |
| CTGF | CCN2 | HCS24 | IGFBP8 | MGC102839 | NOV2 | connective tissue growth factor | - | HPRD | 14585398 |
| CTSG | CG | MGC23078 | cathepsin G | LRP interacts with Cathepsin G. | BIND | 15131125 |
| DAB1 | - | disabled homolog 1 (Drosophila) | - | HPRD,BioGRID | 10827173 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | Reconstituted Complex | BioGRID | 11247302 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 10827173 |
| ELA2 | GE | HLE | HNE | NE | PMN-E | elastase 2, neutrophil | LRP interacts with Elastase 2. | BIND | 15131125 |
| F8 | AHF | DXS1253E | F8B | F8C | FVIII | HEMA | coagulation factor VIII, procoagulant component | - | HPRD,BioGRID | 12522143 |
| F9 | FIX | HEMB | MGC129641 | MGC129642 | PTC | coagulation factor IX | - | HPRD,BioGRID | 12522212 |
| FURIN | FUR | PACE | PCSK3 | SPC1 | furin (paired basic amino acid cleaving enzyme) | - | HPRD | 8546712 |
| GIPC1 | C19orf3 | GIPC | GLUT1CBP | Hs.6454 | IIP-1 | MGC15889 | MGC3774 | NIP | RGS19IP1 | SEMCAP | SYNECTIIN | SYNECTIN | TIP-2 | GIPC PDZ domain containing family, member 1 | Reconstituted Complex Two-hybrid | BioGRID | 10827173 |
| GTPBP1 | GP-1 | GP1 | HSPC018 | MGC20069 | GTP binding protein 1 | - | HPRD | 9820815 |
| GULP1 | CED-6 | CED6 | FLJ31156 | GULP | GULP, engulfment adaptor PTB domain containing 1 | - | HPRD,BioGRID | 11729193 |
| HSP90B1 | ECGP | GP96 | GRP94 | TRA1 | heat shock protein 90kDa beta (Grp94), member 1 | - | HPRD | 11861214 |
| ITGB1BP1 | DKFZp686K08158 | ICAP-1A | ICAP-1B | ICAP-1alpha | ICAP1 | ICAP1A | ICAP1B | integrin beta 1 binding protein 1 | - | HPRD,BioGRID | 10827173 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | - | HPRD,BioGRID | 12888553 |
| LIPC | HDLCQ12 | HL | HTGL | LIPH | lipase, hepatic | - | HPRD,BioGRID | 7721852 |
| LPL | HDLCQ11 | LIPD | lipoprotein lipase | - | HPRD | 1281473|7510694 |7989348 |
| LPL | HDLCQ11 | LIPD | lipoprotein lipase | Reconstituted Complex | BioGRID | 1281473 |7510694 |7989348 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | - | HPRD,BioGRID | 1374383 |1718973 |10747918 |
| LTF | GIG12 | HLF2 | LF | lactotransferrin | - | HPRD | 1464627 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | - | HPRD,BioGRID | 10827173 |
| MAPK8IP2 | IB2 | JIP2 | PRKM8IPL | mitogen-activated protein kinase 8 interacting protein 2 | Reconstituted Complex Two-hybrid | BioGRID | 10827173 |
| MDK | FLJ27379 | MK | NEGF2 | midkine (neurite growth-promoting factor 2) | - | HPRD,BioGRID | 10772929 |
| MMP13 | CLG3 | matrix metallopeptidase 13 (collagenase 3) | - | HPRD | 10514495 |
| MMP14 | MMP-X1 | MT1-MMP | MTMMP1 | matrix metallopeptidase 14 (membrane-inserted) | - | HPRD,BioGRID | 14645246 |
| MMP15 | MT2-MMP | MTMMP2 | SMCP-2 | matrix metallopeptidase 15 (membrane-inserted) | - | HPRD | 14645246 |
| MMP16 | MMP-X2 | MT-MMP2 | MT-MMP3 | MT3-MMP | matrix metallopeptidase 16 (membrane-inserted) | - | HPRD | 14645246 |
| MMP17 | MT4-MMP | matrix metallopeptidase 17 (membrane-inserted) | - | HPRD | 14645246 |
| NOS1AP | 6330408P19Rik | CAPON | MGC138500 | nitric oxide synthase 1 (neuronal) adaptor protein | - | HPRD,BioGRID | 10827173 |
| PDGFB | FLJ12858 | PDGF2 | SIS | SSV | c-sis | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | - | HPRD,BioGRID | 11854294 |
| PDIA2 | PDA2 | PDI | PDIP | PDIR | protein disulfide isomerase family A, member 2 | Affinity Capture-Western | BioGRID | 14980518 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | - | HPRD,BioGRID | 1502153 |10632583 |
| PLAUR | CD87 | UPAR | URKR | plasminogen activator, urokinase receptor | - | HPRD,BioGRID | 11359936 |
| SCN3A | KIAA1356 | NAC3 | Nav1.3 | sodium channel, voltage-gated, type III, alpha subunit | Two-hybrid | BioGRID | 10827173 |
| SERPINA1 | A1A | A1AT | AAT | MGC23330 | MGC9222 | PI | PI1 | PRO2275 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | LRP interacts with alpha-1-PI. | BIND | 15131125 |
| SERPINA3 | AACT | ACT | GIG24 | GIG25 | MGC88254 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | LRP interacts with alpha-1-ACT. | BIND | 15131125 |
| SERPINE1 | PAI | PAI-1 | PAI1 | PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | - | HPRD,BioGRID | 15001579 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11854294 |12789267 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD | 12069755 |12789267 |
| SNX17 | KIAA0064 | sorting nexin 17 | Reconstituted Complex | BioGRID | 12169628 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Biochemical Activity | BioGRID | 11854294 |
| SYNJ2BP | ARIP2 | FLJ11271 | FLJ41973 | OMP25 | synaptojanin 2 binding protein | - | HPRD,BioGRID | 10827173 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD,BioGRID | 9045712 |14991768 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| ST WNT BETA CATENIN PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| JOHANSSON BRAIN CANCER EARLY VS LATE DN | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY UP | 61 | 34 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC | 50 | 32 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C2 | 25 | 18 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| ABE INNER EAR | 48 | 26 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| MIZUKAMI HYPOXIA DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 213 | 219 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-200bc/429 | 167 | 174 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-205 | 118 | 125 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-369-3p | 169 | 175 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-496 | 110 | 116 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.