Gene Page: SMAD4
Summary ?
| GeneID | 4089 |
| Symbol | SMAD4 |
| Synonyms | DPC4|JIP|MADH4|MYHRS |
| Description | SMAD family member 4 |
| Reference | MIM:600993|HGNC:HGNC:6770|Ensembl:ENSG00000141646|HPRD:02995|Vega:OTTHUMG00000132696 |
| Gene type | protein-coding |
| Map location | 18q21.1 |
| Pascal p-value | 0.201 |
| Sherlock p-value | 0.119 |
| Fetal beta | 1.113 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 4 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 4 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0482 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10315128 | 18 | 48556631 | SMAD4 | -0.021 | 0.35 | DMG:Nishioka_2013 | |
| cg27644254 | 18 | 48556578 | SMAD4 | 4.17E-8 | -0.008 | 1.16E-5 | DMG:Jaffe_2016 |
| cg23885062 | 18 | 48556580 | SMAD4 | 8.29E-8 | -0.008 | 1.93E-5 | DMG:Jaffe_2016 |
| cg10315128 | 18 | 48556631 | SMAD4 | 9.67E-8 | -0.006 | 2.16E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
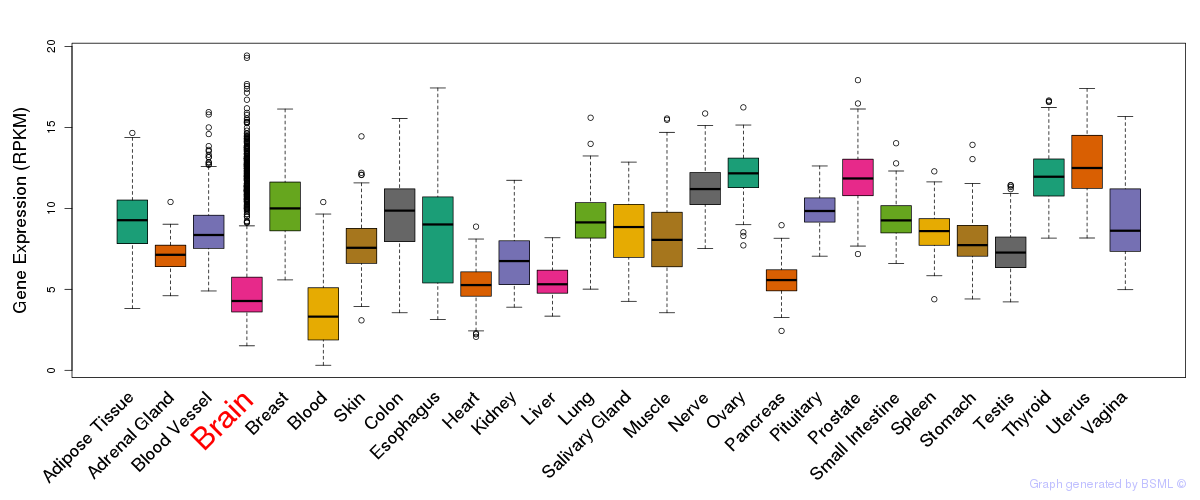
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0016563 | transcription activator activity | IEA | - | |
| GO:0042803 | protein homodimerization activity | IPI | 8774881 | |
| GO:0043565 | sequence-specific DNA binding | IDA | 10823886 | |
| GO:0046332 | SMAD binding | IPI | 8774881 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001666 | response to hypoxia | IMP | 12411310 | |
| GO:0001658 | ureteric bud branching | IEA | - | |
| GO:0001702 | gastrulation with mouth forming second | IEA | - | |
| GO:0001822 | kidney development | IEA | - | |
| GO:0007183 | SMAD protein complex assembly | IDA | 10823886 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | IMP | 8774881 | |
| GO:0051098 | regulation of binding | IEA | - | |
| GO:0030509 | BMP signaling pathway | EXP | 15621726 | |
| GO:0032909 | regulation of transforming growth factor-beta2 production | IMP | 12411310 | |
| GO:0030308 | negative regulation of cell growth | IDA | 8774881 | |
| GO:0045892 | negative regulation of transcription, DNA-dependent | IDA | 8774881 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9311995 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 10549282 |11121043 | |
| GO:0005667 | transcription factor complex | IPI | 12374795 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVR1B | ACTRIB | ACVRLK4 | ALK4 | SKR2 | activin A receptor, type IB | - | HPRD | 9892009 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Affinity Capture-Western | BioGRID | 16362038 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 12226080 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | - | HPRD,BioGRID | 10085140 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 interacts with Smad4. This interaction was modeled on a demonstrated interaction between BRCA1 from human and Smad4 from an unspecified species. | BIND | 15735739 |
| BTRC | BETA-TRCP | FBW1A | FBXW1 | FBXW1A | FWD1 | MGC4643 | bTrCP | bTrCP1 | betaTrCP | beta-transducin repeat containing | - | HPRD,BioGRID | 14988407 |
| CAMK2G | CAMK | CAMK-II | CAMKG | FLJ16043 | MGC26678 | calcium/calmodulin-dependent protein kinase II gamma | - | HPRD,BioGRID | 11027280 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Smad4 binds the distal region of the p21Cip1 promoter. | BIND | 15084259 |
| CEBPA | C/EBP-alpha | CEBP | CCAAT/enhancer binding protein (C/EBP), alpha | Reconstituted Complex | BioGRID | 12524424 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | CEBPB (C/EBP-beta) interacts with SMAD4. This interaction was modeled on a demonstrated interaction between CEBPB and SMAD4, both from unspecified species. | BIND | 12202226 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 12524424 |
| CEBPD | C/EBP-delta | CELF | CRP3 | NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta | Affinity Capture-Western Reconstituted Complex | BioGRID | 12524424 |
| CITED1 | MSG1 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 | - | HPRD,BioGRID | 9707553 |
| COPS5 | CSN5 | JAB1 | MGC3149 | MOV-34 | SGN5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | - | HPRD,BioGRID | 11818334 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD,BioGRID | 12000714 |
| DACH1 | DACH | FLJ10138 | dachshund homolog 1 (Drosophila) | - | HPRD | 14525983 |
| DCP1A | FLJ21691 | HSA275986 | Nbla00360 | SMAD4IP1 | SMIF | DCP1 decapping enzyme homolog A (S. cerevisiae) | - | HPRD,BioGRID | 11836524 |
| DLX1 | - | distal-less homeobox 1 | - | HPRD,BioGRID | 14671321 |
| DVL1 | DVL | MGC54245 | dishevelled, dsh homolog 1 (Drosophila) | Reconstituted Complex | BioGRID | 12650946 |
| ECSIT | SITPEC | ECSIT homolog (Drosophila) | - | HPRD | 14633973 |
| EID2 | CRI2 | EID-2 | MGC20452 | EP300 interacting inhibitor of differentiation 2 | - | HPRD,BioGRID | 14612439 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western | BioGRID | 10887155 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p300 interacts with Smad4 fragment. p300 interacting with full-length Smad4 could not be demonstrated in vitro. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human Smad4 fragment. | BIND | 9679056 |
| ERBB2IP | ERBIN | LAP2 | erbb2 interacting protein | - | HPRD,BioGRID | 12650946 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Two-hybrid | BioGRID | 16189514 |
| FOXH1 | FAST-1 | FAST1 | forkhead box H1 | Reconstituted Complex Two-hybrid | BioGRID | 9288972 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | FoxO1 interacts with Smad4. | BIND | 15084259 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | - | HPRD,BioGRID | 15084259 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | - | HPRD,BioGRID | 15084259 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | FoxO3 interacts with Smad4. | BIND | 15084259 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | - | HPRD,BioGRID | 15084259 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | FoxO4 interacts with Smad4. | BIND | 15084259 |
| FSTL3 | FLRG | FSRP | follistatin-like 3 (secreted glycoprotein) | - | HPRD | 11571638 |
| GLI3 | ACLS | GCPS | PAP-A | PAPA | PAPA1 | PAPB | PHS | PPDIV | GLI-Kruppel family member GLI3 | - | HPRD | 9843199 |
| HOXA9 | ABD-B | HOX1 | HOX1.7 | HOX1G | MGC1934 | homeobox A9 | - | HPRD,BioGRID | 11042172 |
| HOXC8 | HOX3 | HOX3A | homeobox C8 | - | HPRD,BioGRID | 10224145 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD | 10220381 |
| JUNB | AP-1 | jun B proto-oncogene | - | HPRD,BioGRID | 10220381 |
| JUND | AP-1 | jun D proto-oncogene | - | HPRD | 10220381 |
| KPNB1 | IMB1 | IPOB | Impnb | MGC2155 | MGC2156 | MGC2157 | NTF97 | karyopherin (importin) beta 1 | Affinity Capture-Western | BioGRID | 10846168 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | - | HPRD,BioGRID | 10890911 |
| MAX | MGC10775 | MGC11225 | MGC18164 | MGC34679 | MGC36767 | bHLHd4 | orf1 | MYC associated factor X | - | HPRD,BioGRID | 12551947 |
| MDM4 | DKFZp781B1423 | HDMX | MDMX | MGC132766 | MRP1 | Mdm4 p53 binding protein homolog (mouse) | Reconstituted Complex | BioGRID | 12483531 |
| MED15 | ARC105 | CAG7A | CTG7A | DKFZp686A2214 | DKFZp762B1216 | FLJ42282 | FLJ42935 | PCQAP | TIG-1 | TIG1 | TNRC7 | mediator complex subunit 15 | - | HPRD | 12167862 |
| MEF2A | ADCAD1 | RSRFC4 | RSRFC9 | myocyte enhancer factor 2A | - | HPRD | 11160896 |
| PARD3B | ALS2CR19 | MGC16131 | PAR3B | PAR3L | PAR3LC | PAR3beta | Par3Lb | par-3 partitioning defective 3 homolog B (C. elegans) | Reconstituted Complex | BioGRID | 12650946 |
| PIAS3 | FLJ14651 | ZMIZ5 | protein inhibitor of activated STAT, 3 | - | HPRD,BioGRID | 14691252 |
| PIAS4 | FLJ12419 | MGC35296 | PIASY | Piasg | ZMIZ6 | protein inhibitor of activated STAT, 4 | Affinity Capture-Western | BioGRID | 12815042 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | - | HPRD,BioGRID | 12150994 |
| SKI | SKV | v-ski sarcoma viral oncogene homolog (avian) | - | HPRD,BioGRID | 12419246 |12857746 |
| SKI | SKV | v-ski sarcoma viral oncogene homolog (avian) | Smad4 interacts with Ski. This interaction was modeled on a demonstrated interaction between human Smad4 and Ski from an unspecified species. | BIND | 15761153 |
| SKI | SKV | v-ski sarcoma viral oncogene homolog (avian) | c-Ski interacts with Smad4. This interaction was modelled on a demonstrated interaction between Smad4 from an unspecified source and human c-Ski. | BIND | 10575014 |
| SKIL | SNO | SnoA | SnoN | SKI-like oncogene | Smad4 interacts with SnoN. This interaction was modeled on a demonstrated interaction between human Smad4 and SnoN from an unspecified species. | BIND | 15761153 |
| SKIL | SNO | SnoA | SnoN | SKI-like oncogene | - | HPRD,BioGRID | 10531062 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | - | HPRD | 10400705 |11700304 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Affinity Capture-Western Co-fractionation | BioGRID | 10531362 |11700304 |12874272 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Smad4 interacts with Smad1. | BIND | 15761153 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 11274206 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Smad4 interacts with Smad2. | BIND | 15761153 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Smad2 interacts with Smad4. This interaction was modeled on a demonstrated interaction between Smad2 from an unspecified species and Smad4 from an unspecified species. | BIND | 9311995 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD | 9311995|9732876 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Smad3 interacts with Smad4. This interaction was modeled on a demonstrated interaction between Smad3 from an unspecified species and Smad4 from an unspecified species. | BIND | 9311995 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Smad4 interacts with Smad3. | BIND | 15761153 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Affinity Capture-Western | BioGRID | 9311995 |9892009 |10531362 |11114293 |12419246 |12794086 |15084259 |
| SMAD5 | DKFZp781C1895 | DKFZp781O1323 | Dwfc | JV5-1 | MADH5 | SMAD family member 5 | Affinity Capture-Western | BioGRID | 10531362 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | Phenotypic Suppression | BioGRID | 9892009 |
| SMAD9 | MADH6 | MADH9 | SMAD8A | SMAD8B | SMAD family member 9 | Affinity Capture-Western Two-hybrid | BioGRID | 10583507 |16189514 |
| SNIP1 | FLJ12553 | RP3-423B22.3 | dJ423B22.2 | Smad nuclear interacting protein 1 | - | HPRD,BioGRID | 10887155 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | Phenotypic Enhancement Two-hybrid | BioGRID | 11278756 |
| SP1 | - | Sp1 transcription factor | - | HPRD | 11432852 |
| SPTBN1 | ELF | SPTB2 | betaSpII | spectrin, beta, non-erythrocytic 1 | - | HPRD | 12543979 |
| STK11IP | KIAA1898 | LIP1 | LKB1IP | STK11IP1 | serine/threonine kinase 11 interacting protein | - | HPRD,BioGRID | 11741830 |
| STUB1 | CHIP | HSPABP2 | NY-CO-7 | SDCCAG7 | UBOX1 | STIP1 homology and U-box containing protein 1 | Smad4 interacts with CHIP. This interaction was modeled on a demonstrated interaction between Smad1 and CHIP, both from an unspecified species. | BIND | 15708501 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | SMAD4 is sumoylated. | BIND | 15592428 |
| TFE3 | RCCP2 | TFEA | bHLHe33 | transcription factor binding to IGHM enhancer 3 | - | HPRD,BioGRID | 10557285 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | ARA55 interacts with Smad4. This interaction was modeled on a demonstrated interaction between human ARA55 and Smad4 from an unspecified species. | BIND | 15561701 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | T-beta-RI phosphorylates hMAD-4. This interaction was modeled on a demonstrated interaction between T-beta-RI from an unspecified species and human hMAD-4. | BIND | 8774881 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | Co-fractionation | BioGRID | 11102446 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | T-beta-RII phosphorylates hMAD-4. This interaction was modeled on a demonstrated interaction between T-beta-RII from an unspecified species and human hMAD-4. | BIND | 8774881 |
| TGFBRAP1 | TRAP-1 | TRAP1 | transforming growth factor, beta receptor associated protein 1 | - | HPRD,BioGRID | 11278302 |
| TOB1 | APRO6 | MGC104792 | MGC34446 | PIG49 | TOB | TROB | TROB1 | transducer of ERBB2, 1 | - | HPRD,BioGRID | 11163184 |
| TRIM33 | FLJ32925 | PTC7 | RFG7 | TF1G | TIF1G | TIF1GAMMA | TIFGAMMA | tripartite motif-containing 33 | Smad4 interacts with Ecto. | BIND | 15820681 |
| TSSK3 | SPOGA3 | STK22C | STK22D | testis-specific serine kinase 3 | SMAD4 interacts with STK22C. This interaction was modeled on a demonstrated interaction between human SMAD4 and mouse STK22C. | BIND | 15761153 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Two-hybrid | BioGRID | 16189514 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD | 12794086 |
| ZNF423 | Ebfaz | KIAA0760 | MGC138520 | MGC138522 | OAZ | Roaz | ZFP423 | Zfp104 | zinc finger protein 423 | - | HPRD,BioGRID | 10660046 |
| ZNF521 | DKFZp564D0764 | EHZF | Evi3 | MGC142182 | MGC142208 | zinc finger protein 521 | - | HPRD | 14630787 |
| ZNF8 | HF.18 | Zfp128 | zinc finger protein 8 | - | HPRD | 12370310 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOB1 PATHWAY | 21 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TGFB PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID ALK2 PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NODAL | 18 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY BMP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK1 AND MAPK2 TARGETS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| DER IFN ALPHA RESPONSE UP | 74 | 48 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-146 | 390 | 397 | 1A,m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-224 | 1194 | 1200 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA | ||||
| miR-26 | 1034 | 1040 | m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-346 | 146 | 152 | 1A | hsa-miR-346brain | UGUCUGCCCGCAUGCCUGCCUCU |
| miR-376c | 966 | 972 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-431 | 1121 | 1127 | m8 | hsa-miR-431 | UGUCUUGCAGGCCGUCAUGCA |
| miR-544 | 982 | 988 | m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.