Gene Page: NF1
Summary ?
| GeneID | 4763 |
| Symbol | NF1 |
| Synonyms | NFNS|VRNF|WSS |
| Description | neurofibromin 1 |
| Reference | MIM:613113|HGNC:HGNC:7765|Ensembl:ENSG00000196712|HPRD:01203|Vega:OTTHUMG00000132871 |
| Gene type | protein-coding |
| Map location | 17q11.2 |
| Pascal p-value | 0.48 |
| Fetal beta | 0.312 |
| DMG | 1 (# studies) |
| Support | STRUCTURAL PLASTICITY G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 12 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0086 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07917842 | 17 | 29672644 | NF1 | 1.1E-5 | 0.488 | 0.013 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
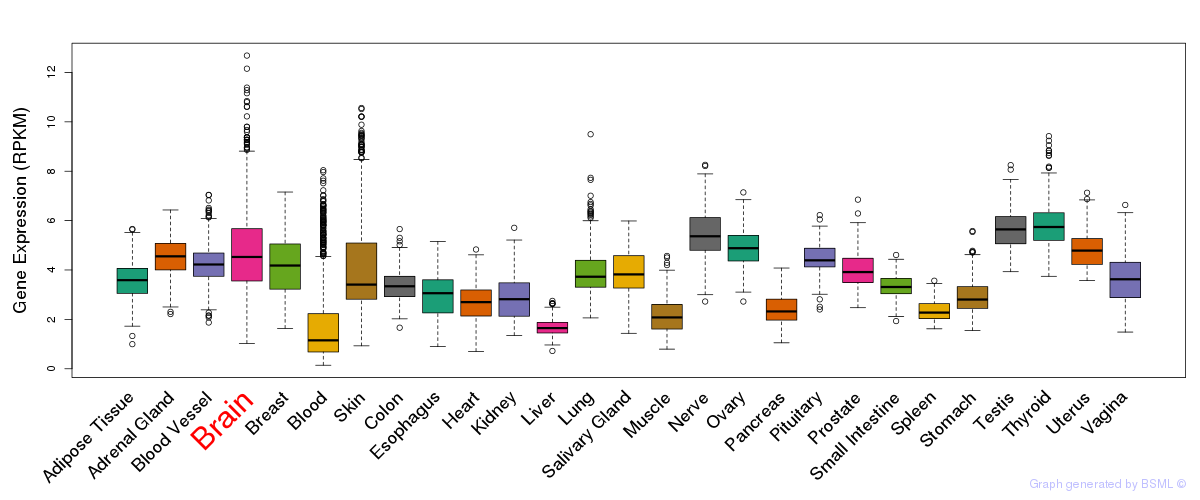
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CRIP2 | 0.88 | 0.88 |
| HTRA2 | 0.87 | 0.86 |
| PIK3R2 | 0.86 | 0.87 |
| C19orf22 | 0.86 | 0.87 |
| CHMP1A | 0.85 | 0.84 |
| UNC119 | 0.85 | 0.85 |
| KLHL22 | 0.85 | 0.86 |
| ADCK2 | 0.85 | 0.84 |
| TBCB | 0.85 | 0.83 |
| TOR3A | 0.84 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.71 | -0.73 |
| AF347015.33 | -0.70 | -0.74 |
| AF347015.8 | -0.70 | -0.74 |
| AF347015.27 | -0.70 | -0.74 |
| MT-CYB | -0.70 | -0.74 |
| AF347015.26 | -0.68 | -0.76 |
| AF347015.31 | -0.68 | -0.71 |
| AF347015.15 | -0.67 | -0.72 |
| AF347015.2 | -0.66 | -0.72 |
| AF347015.21 | -0.63 | -0.70 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0005099 | Ras GTPase activator activity | IDA | 1568247 |1570015 | |
| GO:0005096 | GTPase activator activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 11356864 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | ISS | Brain (GO term level: 7) | - |
| GO:0007406 | negative regulation of neuroblast proliferation | ISS | neurogenesis (GO term level: 10) | - |
| GO:0022011 | myelination in the peripheral nervous system | ISS | neuron, axon, dendrite (GO term level: 14) | - |
| GO:0048715 | negative regulation of oligodendrocyte differentiation | ISS | oligodendrocyte, Glial (GO term level: 12) | - |
| GO:0021897 | forebrain astrocyte development | ISS | neuron, astrocyte, Brain, Glial (GO term level: 11) | - |
| GO:0048485 | sympathetic nervous system development | ISS | neuron, Neurotransmitter (GO term level: 7) | - |
| GO:0045685 | regulation of glial cell differentiation | ISS | Glial (GO term level: 10) | - |
| GO:0021510 | spinal cord development | ISS | Brain (GO term level: 7) | - |
| GO:0048853 | forebrain morphogenesis | ISS | Brain (GO term level: 9) | - |
| GO:0043525 | positive regulation of neuron apoptosis | ISS | neuron (GO term level: 9) | - |
| GO:0001666 | response to hypoxia | ISS | - | |
| GO:0001656 | metanephros development | ISS | - | |
| GO:0001649 | osteoblast differentiation | ISS | - | |
| GO:0001889 | liver development | ISS | - | |
| GO:0001937 | negative regulation of endothelial cell proliferation | IMP | 17404841 | |
| GO:0001952 | regulation of cell-matrix adhesion | ISS | - | |
| GO:0042992 | negative regulation of transcription factor import into nucleus | ISS | - | |
| GO:0007265 | Ras protein signal transduction | ISS | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008542 | visual learning | ISS | - | |
| GO:0008150 | biological_process | ND | - | |
| GO:0007507 | heart development | ISS | - | |
| GO:0007422 | peripheral nervous system development | ISS | - | |
| GO:0014065 | phosphoinositide 3-kinase cascade | ISS | - | |
| GO:0042060 | wound healing | ISS | - | |
| GO:0030036 | actin cytoskeleton organization | ISS | - | |
| GO:0051056 | regulation of small GTPase mediated signal transduction | IEA | - | |
| GO:0021987 | cerebral cortex development | ISS | - | |
| GO:0043473 | pigmentation | ISS | - | |
| GO:0030199 | collagen fibril organization | ISS | - | |
| GO:0032320 | positive regulation of Ras GTPase activity | ISS | - | |
| GO:0030336 | negative regulation of cell migration | IMP | 16648142 | |
| GO:0030325 | adrenal gland development | ISS | - | |
| GO:0043409 | negative regulation of MAPKKK cascade | ISS | - | |
| GO:0048745 | smooth muscle development | ISS | - | |
| GO:0043407 | negative regulation of MAP kinase activity | ISS | - | |
| GO:0045762 | positive regulation of adenylate cyclase activity | ISS | - | |
| GO:0045765 | regulation of angiogenesis | IMP | 17404841 | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0045124 | regulation of bone resorption | ISS | - | |
| GO:0043535 | regulation of blood vessel endothelial cell migration | IMP | 17404841 | |
| GO:0048593 | camera-type eye morphogenesis | ISS | - | |
| GO:0050890 | cognition | IMP | 17299016 | |
| GO:0048844 | artery morphogenesis | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030425 | dendrite | IDA | neuron, axon, dendrite (GO term level: 6) | 1550670 |
| GO:0030424 | axon | IDA | neuron, axon, Neurotransmitter (GO term level: 6) | 1550670 |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | ISS | 1550670 | |
| GO:0005737 | cytoplasm | ISS | 1550670 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID RAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 AND DCC TARGETS | 35 | 25 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 12 | 11 | 8 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| COURTOIS SENESCENCE TRIGGERS | 6 | 5 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED FREQUENTLY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 UP | 45 | 32 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA MUTATED | 8 | 8 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 914 | 920 | 1A | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-103/107 | 177 | 183 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA | ||||
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-128 | 1195 | 1202 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-137 | 650 | 656 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-153 | 2974 | 2980 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-182 | 1733 | 1740 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-27 | 1196 | 1203 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 1702 | 1709 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-369-3p | 3119 | 3125 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-370 | 3358 | 3365 | 1A,m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-378 | 267 | 274 | 1A,m8 | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-381 | 3264 | 3270 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-448 | 2974 | 2980 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-485-5p | 540 | 546 | 1A | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-486 | 659 | 665 | 1A | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-490 | 1435 | 1441 | m8 | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
| miR-496 | 1645 | 1651 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
| miR-543 | 2969 | 2975 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-96 | 1734 | 1740 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.