Gene Page: NFYA
Summary ?
| GeneID | 4800 |
| Symbol | NFYA |
| Synonyms | CBF-A|CBF-B|HAP2|NF-YA |
| Description | nuclear transcription factor Y subunit alpha |
| Reference | MIM:189903|HGNC:HGNC:7804|HPRD:01793| |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 0.497 |
| Fetal beta | 1.563 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12694820 | 6 | 41041189 | NFYA | 1.6E-8 | -0.01 | 5.94E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
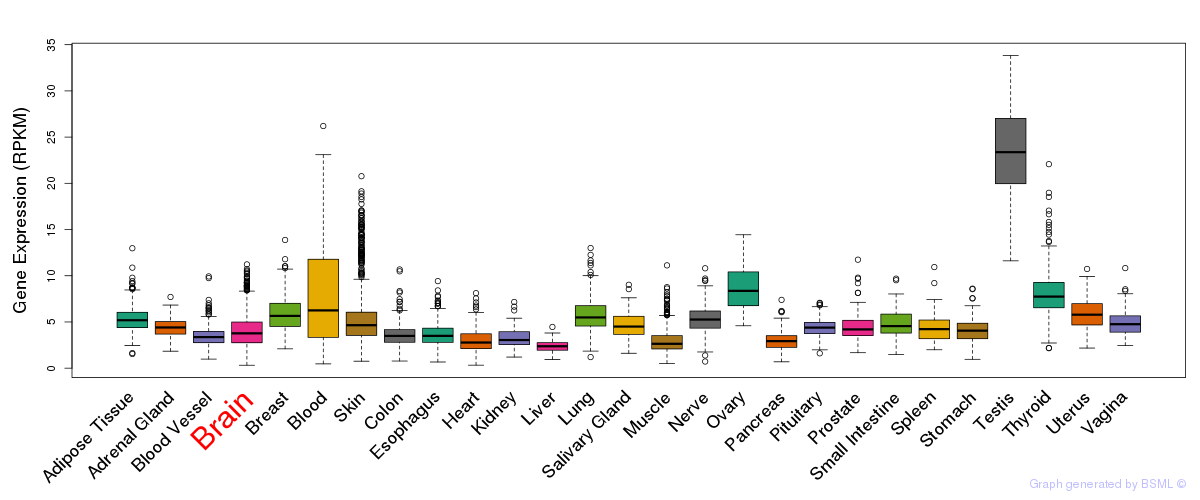
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NME1-NME2 | 0.98 | 0.98 |
| C2orf79 | 0.89 | 0.91 |
| MYL6 | 0.89 | 0.89 |
| NME2P1 | 0.89 | 0.84 |
| RPL12 | 0.89 | 0.83 |
| COMMD6 | 0.86 | 0.84 |
| RPL36 | 0.86 | 0.81 |
| TCTEX1D2 | 0.85 | 0.84 |
| RP11-544M22.1 | 0.85 | 0.83 |
| UFC1 | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MYH14 | -0.53 | -0.65 |
| AF347015.26 | -0.53 | -0.65 |
| AF347015.33 | -0.51 | -0.61 |
| EPAS1 | -0.51 | -0.61 |
| KIF1C | -0.51 | -0.65 |
| ZBTB7B | -0.51 | -0.62 |
| AF347015.15 | -0.51 | -0.60 |
| AF347015.27 | -0.50 | -0.59 |
| FYCO1 | -0.50 | -0.61 |
| AF347015.8 | -0.50 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 1774067 |2000400 | |
| GO:0005515 | protein binding | IPI | 10571058 |12659632 |12741956 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IDA | 15243141 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 2000400 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 15243141 | |
| GO:0016602 | CCAAT-binding factor complex | IDA | 15243141 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APPBP2 | HS.84084 | KIAA0228 | PAT1 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| ATF6 | ATF6A | activating transcription factor 6 | Two-hybrid | BioGRID | 11158310 |
| ATF6B | CREB-RP | CREBL1 | FLJ10066 | G13 | activating transcription factor 6 beta | Two-hybrid | BioGRID | 11158310 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 11777930 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 interacts with NF-YA | BIND | 11777930 |
| ELF1 | - | E74-like factor 1 (ets domain transcription factor) | Reconstituted Complex | BioGRID | 9668064 |
| HMGA1 | HMG-R | HMGA1A | HMGIY | MGC12816 | MGC4242 | MGC4854 | high mobility group AT-hook 1 | - | HPRD,BioGRID | 9388234 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | Affinity Capture-Western | BioGRID | 11903046 |
| NFYB | CBF-A | CBF-B | HAP3 | NF-YB | nuclear transcription factor Y, beta | - | HPRD,BioGRID | 8051128 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | - | HPRD | 11777930 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Oct-1 interacts with NF-YA | BIND | 11777930 |
| SPI1 | OF | PU.1 | SFPI1 | SPI-1 | SPI-A | spleen focus forming virus (SFFV) proviral integration oncogene spi1 | Reconstituted Complex | BioGRID | 9668064 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | NF-YA interacts with SRF. This interaction was modeled on a demonstrated interaction between NF-YA from an unspecified species and SRF from human. | BIND | 10571058 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | Reconstituted Complex Two-hybrid | BioGRID | 10571058 |
| ZHX1 | - | zinc fingers and homeoboxes 1 | NF-YA interacts with ZHX1. This interaction was modeled on a demonstrated interaction between NF-YA from an unspecified species and ZHX1 from human. | BIND | 10571058 |
| ZHX1 | - | zinc fingers and homeoboxes 1 | - | HPRD,BioGRID | 10441475 |10571058 |
| ZHX2 | AFR1 | KIAA0854 | RAF | zinc fingers and homeoboxes 2 | - | HPRD | 12741956 |
| ZHX3 | KIAA0395 | TIX1 | zinc fingers and homeoboxes 3 | - | HPRD | 12659632 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PERK REGULATED GENE EXPRESSION | 29 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF GENES BY ATF4 | 26 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREEN DN | 25 | 12 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| ADDYA ERYTHROID DIFFERENTIATION BY HEMIN | 73 | 47 | All SZGR 2.0 genes in this pathway |
| KANG FLUOROURACIL RESISTANCE DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C CLUSTER UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C UP | 47 | 29 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-132/212 | 221 | 227 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-200bc/429 | 142 | 149 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-22 | 1274 | 1280 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-330 | 686 | 692 | 1A | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-504 | 1390 | 1397 | 1A,m8 | hsa-miR-504 | AGACCCUGGUCUGCACUCUAU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 294 | 301 | 1A,m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.