Gene Page: NTRK2
Summary ?
| GeneID | 4915 |
| Symbol | NTRK2 |
| Synonyms | GP145-TrkB|TRKB|trk-B |
| Description | neurotrophic tyrosine kinase, receptor, type 2 |
| Reference | MIM:600456|HGNC:HGNC:8032|Ensembl:ENSG00000148053|HPRD:02712|Vega:OTTHUMG00000020120 |
| Gene type | protein-coding |
| Map location | 9q22.1 |
| Pascal p-value | 0.194 |
| Fetal beta | -1.567 |
| DMG | 1 (# studies) |
| Support | TYROSINE KINASE SIGNALING G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03628748 | 9 | 87285133 | NTRK2 | 4.08E-8 | -0.022 | 1.15E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
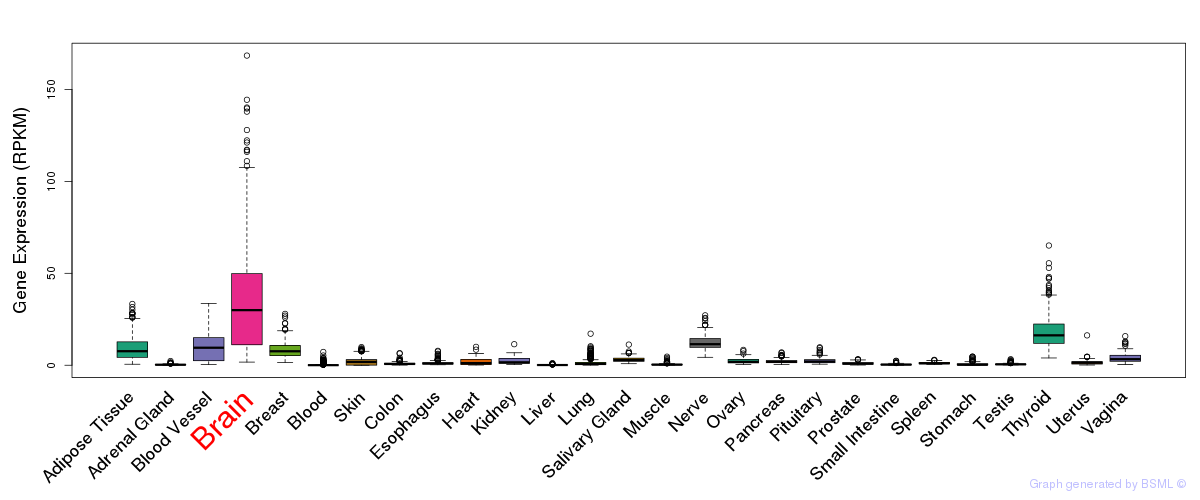
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACOT8 | 0.94 | 0.90 |
| TUSC2 | 0.88 | 0.86 |
| MECR | 0.88 | 0.88 |
| UROD | 0.88 | 0.84 |
| MRPL17 | 0.87 | 0.88 |
| MRPL2 | 0.87 | 0.85 |
| SDHB | 0.87 | 0.88 |
| C9orf123 | 0.87 | 0.83 |
| CDK5 | 0.87 | 0.84 |
| COX4I1 | 0.87 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.50 | -0.58 |
| AC010300.1 | -0.47 | -0.56 |
| AC005921.3 | -0.46 | -0.65 |
| SFRS18 | -0.45 | -0.59 |
| ZNF814 | -0.44 | -0.53 |
| ANKRD36B | -0.42 | -0.57 |
| ANKRD36 | -0.41 | -0.54 |
| IL3RA | -0.41 | -0.58 |
| RBM25 | -0.41 | -0.38 |
| AF347015.18 | -0.40 | -0.18 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | IEA | neurite (GO term level: 8) | - |
| GO:0043121 | neurotrophin binding | TAS | neuron (GO term level: 5) | 1710174 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0007190 | activation of adenylate cyclase activity | EXP | 16805430 | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | EXP | 16939974 | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IEA | - | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | TAS | 1710174 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1710174 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BDNF | MGC34632 | brain-derived neurotrophic factor | - | HPRD,BioGRID | 9312147 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD,BioGRID | 11157096 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | Reconstituted Complex | BioGRID | 10092678 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 9648856 |
| GIPC1 | C19orf3 | GIPC | GLUT1CBP | Hs.6454 | IIP-1 | MGC15889 | MGC3774 | NIP | RGS19IP1 | SEMCAP | SYNECTIIN | SYNECTIN | TIP-2 | GIPC PDZ domain containing family, member 1 | - | HPRD,BioGRID | 11251075 |
| KIDINS220 | ARMS | MGC163482 | kinase D-interacting substrate, 220kDa | - | HPRD | 15167895 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD | 12074588 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD,BioGRID | 12074588 |
| NTF3 | HDNF | MGC129711 | NGF-2 | NGF2 | NT3 | neurotrophin 3 | - | HPRD,BioGRID | 8621424 |
| NTF4 | NT-4/5 | NT4 | NT5 | NTF5 | neurotrophin 4 | Interaction between TRKB (PDB ID: 1HCF_Y) and NTF4 (PDB ID: 1HCF_A). | BIND | 10388563 |10490030 |10631974 |11263982 |11738045 |
| NTF4 | NT-4/5 | NT4 | NT5 | NTF5 | neurotrophin 4 | Interaction between TRKB (PDB ID: 1HCF_X) and NTF4 (PDB ID: 1HCF_B). | BIND | 10388563 |10490030 |10631974 |11263982 |11738045 |
| NTF4 | NT-4/5 | NT4 | NT5 | NTF5 | neurotrophin 4 | - | HPRD,BioGRID | 11855816 |
| NTF4 | NT-4/5 | NT4 | NT5 | NTF5 | neurotrophin 4 | Interaction between TRKB (PDB ID: 1HCF_Y) and NTF4 (PDB ID: 1HCF_B). | BIND | 10388563 |10490030 |10631974 |11263982 |11738045 |
| NTF4 | NT-4/5 | NT4 | NT5 | NTF5 | neurotrophin 4 | Interaction between TRKB (PDB ID: 1HCF_X) and NTF4 (PDB ID: 1HCF_A). | BIND | 10388563 |10490030 |10631974 |11263982 |11738045 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Two-hybrid | BioGRID | 12074588 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Reconstituted Complex Two-hybrid | BioGRID | 10092678 |12074588 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 12237455 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | Two-hybrid | BioGRID | 12074588 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | - | HPRD | 9856458 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Reconstituted Complex | BioGRID | 10092678 |
| SHC3 | N-Shc | NSHC | RAI | SHCC | SHC (Src homology 2 domain containing) transforming protein 3 | - | HPRD,BioGRID | 9507002 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | - | HPRD,BioGRID | 12471037 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS UP | 77 | 62 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS DN | 24 | 13 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MULTIPLE MYELOMA WITH 14Q32 TRANSLOCATIONS | 36 | 25 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR DN | 57 | 45 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MF UP | 47 | 27 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| HUNSBERGER EXERCISE REGULATED GENES | 31 | 26 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER BY MUTATION RATE | 20 | 18 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-138 | 507 | 513 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.