Gene Page: TNFRSF11B
Summary ?
| GeneID | 4982 |
| Symbol | TNFRSF11B |
| Synonyms | OCIF|OPG|PDB5|TR1 |
| Description | tumor necrosis factor receptor superfamily member 11b |
| Reference | MIM:602643|HGNC:HGNC:11909|Ensembl:ENSG00000164761|HPRD:04032|Vega:OTTHUMG00000164969 |
| Gene type | protein-coding |
| Map location | 8q24 |
| Pascal p-value | 0.197 |
| Fetal beta | -0.652 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 3 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs6993813 | 8 | 120052238 | null | 0.6515 | TNFRSF11B | null | |
| rs4355801 | 8 | 119923873 | null | 0.6482 | TNFRSF11B | null | |
| rs6469804 | 8 | 120044829 | null | 0.6666 | TNFRSF11B | null |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | TNFRSF11B | 4982 | 2.161E-4 | trans | ||
| rs546975 | chr1 | 91963738 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs17535450 | chr2 | 39900012 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs272110 | chr2 | 131546319 | TNFRSF11B | 4982 | 0.18 | trans | ||
| rs9814206 | chr3 | 74573226 | TNFRSF11B | 4982 | 0.11 | trans | ||
| rs1691463 | chr3 | 84914366 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs394185 | chr3 | 84959076 | TNFRSF11B | 4982 | 0.09 | trans | ||
| rs9288980 | chr3 | 112964991 | TNFRSF11B | 4982 | 0.16 | trans | ||
| rs3995908 | chr3 | 112998529 | TNFRSF11B | 4982 | 0.16 | trans | ||
| rs7652801 | chr3 | 113008605 | TNFRSF11B | 4982 | 0.16 | trans | ||
| rs2922180 | chr3 | 125280472 | TNFRSF11B | 4982 | 7.331E-10 | trans | ||
| rs2971271 | chr3 | 125450402 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs2976809 | chr3 | 125451738 | TNFRSF11B | 4982 | 2.874E-6 | trans | ||
| rs9917710 | chr3 | 176280218 | TNFRSF11B | 4982 | 0.18 | trans | ||
| rs16889842 | chr4 | 9872012 | TNFRSF11B | 4982 | 0.03 | trans | ||
| snp_a-1976228 | 0 | TNFRSF11B | 4982 | 0.13 | trans | |||
| snp_a-2065799 | 0 | TNFRSF11B | 4982 | 0.06 | trans | |||
| rs6856952 | chr4 | 74394402 | TNFRSF11B | 4982 | 0.06 | trans | ||
| rs17005214 | chr4 | 82845432 | TNFRSF11B | 4982 | 0.06 | trans | ||
| rs2732167 | chr4 | 88706292 | TNFRSF11B | 4982 | 0.17 | trans | ||
| rs313946 | chr4 | 113975904 | TNFRSF11B | 4982 | 0.07 | trans | ||
| rs17034860 | chr4 | 135177933 | TNFRSF11B | 4982 | 0.14 | trans | ||
| rs17636193 | chr5 | 3540160 | TNFRSF11B | 4982 | 0.09 | trans | ||
| rs1867710 | chr5 | 3576228 | TNFRSF11B | 4982 | 0.15 | trans | ||
| rs11747698 | chr5 | 3577007 | TNFRSF11B | 4982 | 0.15 | trans | ||
| rs13174189 | chr5 | 3579173 | TNFRSF11B | 4982 | 0.19 | trans | ||
| rs13187916 | chr5 | 3580498 | TNFRSF11B | 4982 | 0.15 | trans | ||
| rs13170862 | chr5 | 3582215 | TNFRSF11B | 4982 | 0.15 | trans | ||
| rs10050805 | chr5 | 30805352 | TNFRSF11B | 4982 | 0 | trans | ||
| rs837115 | chr5 | 40736290 | TNFRSF11B | 4982 | 0.03 | trans | ||
| rs16894149 | chr5 | 61013724 | TNFRSF11B | 4982 | 0.1 | trans | ||
| rs17143695 | chr5 | 117520843 | TNFRSF11B | 4982 | 0.06 | trans | ||
| rs11954814 | chr5 | 155088338 | TNFRSF11B | 4982 | 0.2 | trans | ||
| rs17056435 | chr5 | 158453782 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs17135501 | chr6 | 2704474 | TNFRSF11B | 4982 | 0.04 | trans | ||
| rs17135521 | chr6 | 2707631 | TNFRSF11B | 4982 | 0.04 | trans | ||
| snp_a-1787926 | 0 | TNFRSF11B | 4982 | 0.04 | trans | |||
| rs6920042 | chr6 | 67907570 | TNFRSF11B | 4982 | 0.14 | trans | ||
| rs6939897 | chr6 | 67913123 | TNFRSF11B | 4982 | 0.14 | trans | ||
| rs6455226 | chr6 | 67930295 | TNFRSF11B | 4982 | 6.269E-4 | trans | ||
| rs2064428 | chr6 | 85067126 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs9451867 | chr6 | 92659609 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs283058 | chr6 | 118625224 | TNFRSF11B | 4982 | 0.14 | trans | ||
| rs2398707 | chr7 | 2113428 | TNFRSF11B | 4982 | 0.03 | trans | ||
| rs3778948 | chr7 | 2123290 | TNFRSF11B | 4982 | 0.09 | trans | ||
| rs3778951 | chr7 | 2125319 | TNFRSF11B | 4982 | 0.03 | trans | ||
| rs3778957 | chr7 | 2127710 | TNFRSF11B | 4982 | 0.03 | trans | ||
| rs3778958 | chr7 | 2127794 | TNFRSF11B | 4982 | 0.09 | trans | ||
| rs3778968 | chr7 | 2139903 | TNFRSF11B | 4982 | 0.09 | trans | ||
| rs2286089 | chr7 | 38789903 | TNFRSF11B | 4982 | 0.11 | trans | ||
| rs10085487 | chr7 | 38870379 | TNFRSF11B | 4982 | 0.06 | trans | ||
| rs7782277 | chr7 | 38938053 | TNFRSF11B | 4982 | 0.16 | trans | ||
| rs2430483 | chr7 | 104548631 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs1960369 | chr8 | 6379235 | TNFRSF11B | 4982 | 0.1 | trans | ||
| rs2199402 | chr8 | 9201002 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs2169384 | chr8 | 9216985 | TNFRSF11B | 4982 | 0.06 | trans | ||
| rs1381350 | chr8 | 9217194 | TNFRSF11B | 4982 | 0.17 | trans | ||
| rs7841407 | chr8 | 9243427 | TNFRSF11B | 4982 | 0 | trans | ||
| rs2739708 | chr8 | 17992280 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs714131 | chr8 | 17992832 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs2739726 | chr8 | 17998576 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs2919377 | chr8 | 32573740 | TNFRSF11B | 4982 | 0.04 | trans | ||
| rs9410375 | chr9 | 91444622 | TNFRSF11B | 4982 | 0.06 | trans | ||
| rs2253777 | chr9 | 93467893 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs4743989 | chr9 | 97859191 | TNFRSF11B | 4982 | 0.13 | trans | ||
| rs998410 | chr9 | 117622673 | TNFRSF11B | 4982 | 0.06 | trans | ||
| rs9423711 | chr10 | 2574802 | TNFRSF11B | 4982 | 0.03 | trans | ||
| rs651134 | chr10 | 27549960 | TNFRSF11B | 4982 | 0.17 | trans | ||
| rs748375 | chr10 | 33440178 | TNFRSF11B | 4982 | 0.07 | trans | ||
| rs4514358 | chr10 | 43861923 | TNFRSF11B | 4982 | 0.05 | trans | ||
| rs7081666 | chr10 | 47673800 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs10886877 | chr10 | 85425690 | TNFRSF11B | 4982 | 0.03 | trans | ||
| rs10488758 | chr11 | 103493095 | TNFRSF11B | 4982 | 0.04 | trans | ||
| rs1905067 | chr11 | 103493989 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs10895512 | chr11 | 103553194 | TNFRSF11B | 4982 | 0.04 | trans | ||
| rs11604394 | chr11 | 103554879 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs11603978 | chr11 | 103555057 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs11225972 | chr11 | 103555992 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs11225974 | chr11 | 103560499 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs11225990 | chr11 | 103608380 | TNFRSF11B | 4982 | 0.04 | trans | ||
| rs10747972 | 0 | TNFRSF11B | 4982 | 0.08 | trans | |||
| rs10879027 | chr12 | 70237156 | TNFRSF11B | 4982 | 2.689E-4 | trans | ||
| rs11112163 | chr12 | 105072508 | TNFRSF11B | 4982 | 0 | trans | ||
| rs9551590 | chr13 | 29589464 | TNFRSF11B | 4982 | 0.05 | trans | ||
| rs17073665 | chr13 | 81541961 | TNFRSF11B | 4982 | 0 | trans | ||
| rs11840833 | chr13 | 94378306 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs16975617 | chr13 | 110867407 | TNFRSF11B | 4982 | 0.09 | trans | ||
| rs10142086 | chr14 | 35135892 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs1951867 | chr14 | 54407191 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs2093759 | chr14 | 97171024 | TNFRSF11B | 4982 | 0 | trans | ||
| rs17244419 | chr14 | 97171074 | TNFRSF11B | 4982 | 5.97E-5 | trans | ||
| rs17629301 | chr14 | 99467034 | TNFRSF11B | 4982 | 0.05 | trans | ||
| rs7177212 | chr15 | 34425413 | TNFRSF11B | 4982 | 0.11 | trans | ||
| rs16976022 | chr15 | 55454260 | TNFRSF11B | 4982 | 0.19 | trans | ||
| rs16976029 | chr15 | 55455268 | TNFRSF11B | 4982 | 0.17 | trans | ||
| rs16976036 | chr15 | 55456921 | TNFRSF11B | 4982 | 0.13 | trans | ||
| rs8034402 | chr15 | 55480932 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs8043029 | chr15 | 55486010 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs3809540 | chr15 | 55489411 | TNFRSF11B | 4982 | 0.02 | trans | ||
| rs7165491 | chr15 | 55500454 | TNFRSF11B | 4982 | 0.18 | trans | ||
| rs7165622 | chr15 | 93979910 | TNFRSF11B | 4982 | 0.19 | trans | ||
| rs4984405 | chr15 | 95260170 | TNFRSF11B | 4982 | 0.14 | trans | ||
| rs13380712 | chr16 | 25601181 | TNFRSF11B | 4982 | 0.05 | trans | ||
| rs16977927 | chr17 | 72154232 | TNFRSF11B | 4982 | 0.08 | trans | ||
| rs7236104 | chr18 | 14032818 | TNFRSF11B | 4982 | 0.18 | trans | ||
| rs4987765 | chr18 | 60905021 | TNFRSF11B | 4982 | 0.12 | trans | ||
| rs11873703 | chr18 | 61448702 | TNFRSF11B | 4982 | 0 | trans | ||
| rs6017989 | chr20 | 45329846 | TNFRSF11B | 4982 | 0.19 | trans | ||
| rs6095741 | chr20 | 48666589 | TNFRSF11B | 4982 | 6E-7 | trans | ||
| rs5991662 | chrX | 43335796 | TNFRSF11B | 4982 | 0.01 | trans | ||
| rs2254586 | chrX | 117855503 | TNFRSF11B | 4982 | 0.14 | trans |
Section II. Transcriptome annotation
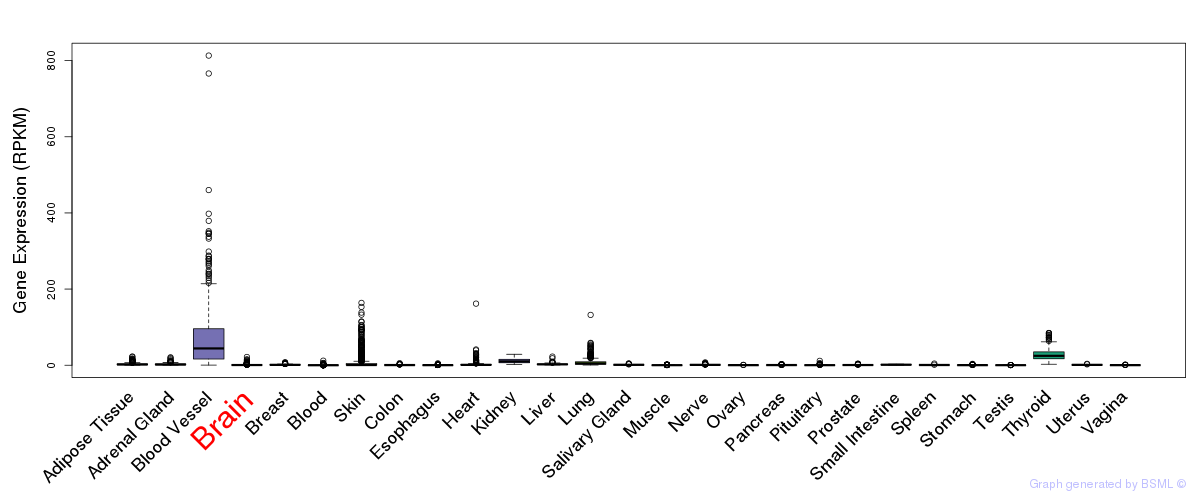
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| PIEPOLI LGI1 TARGETS UP | 15 | 11 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 4NM UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 1 UP | 36 | 18 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 SIGNALING 2 | 12 | 8 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 TARGETS WITH STAT5 BINDING SITES T1 | 9 | 7 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS SUBSET | 33 | 20 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| WILLERT WNT SIGNALING | 24 | 13 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| LY AGING MIDDLE UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO RADIATION THERAPY | 32 | 20 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY DN | 38 | 27 | All SZGR 2.0 genes in this pathway |
| HOFFMAN CLOCK TARGETS UP | 11 | 7 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| PLASARI NFIC TARGETS BASAL UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS UP | 48 | 33 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |