Gene Page: PCSK5
Summary ?
| GeneID | 5125 |
| Symbol | PCSK5 |
| Synonyms | PC5|PC6|PC6A|SPC6 |
| Description | proprotein convertase subtilisin/kexin type 5 |
| Reference | MIM:600488|HGNC:HGNC:8747|Ensembl:ENSG00000099139|HPRD:08985|Vega:OTTHUMG00000020039 |
| Gene type | protein-coding |
| Map location | 9q21.3 |
| Pascal p-value | 0.088 |
| Sherlock p-value | 0.26 |
| Fetal beta | 1.189 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16904306 | 9 | 78506294 | PCSK5 | -0.018 | 0.68 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4549432 | chr4 | 182350656 | PCSK5 | 5125 | 0.08 | trans |
Section II. Transcriptome annotation
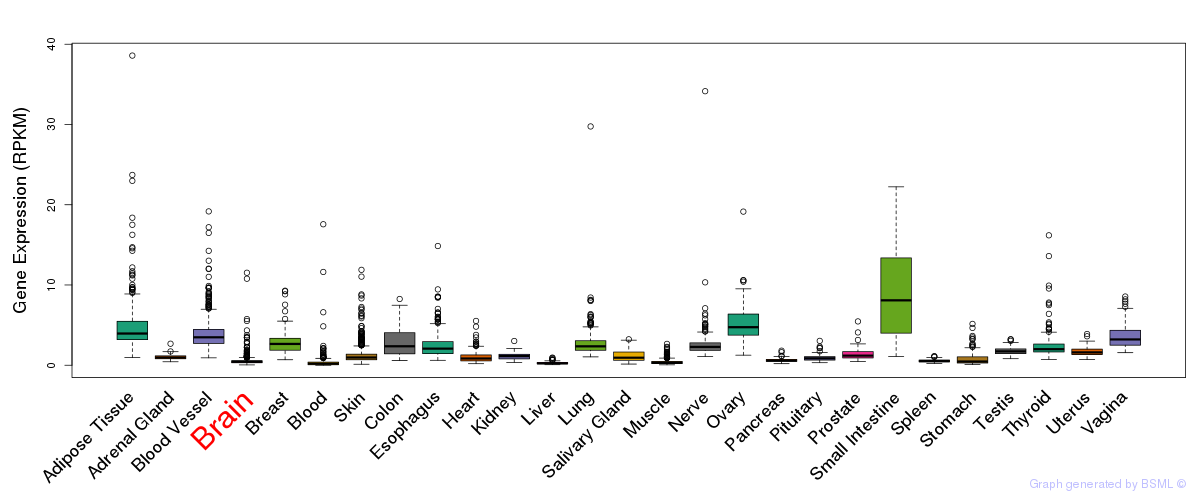
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TUBA8 | 0.60 | 0.63 |
| DTNBP1 | 0.59 | 0.47 |
| PCP4 | 0.58 | 0.49 |
| ECE2 | 0.57 | 0.59 |
| BSPRY | 0.57 | 0.62 |
| IKBKE | 0.57 | 0.44 |
| KCTD17 | 0.56 | 0.38 |
| HRH3 | 0.56 | 0.57 |
| TMEM173 | 0.55 | 0.46 |
| CYB5R1 | 0.55 | 0.46 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH2B2 | -0.36 | -0.46 |
| SOX12 | -0.35 | -0.39 |
| PFN3 | -0.34 | -0.41 |
| MEX3D | -0.33 | -0.36 |
| DSCC1 | -0.33 | -0.38 |
| FOXO6 | -0.33 | -0.35 |
| LENG9 | -0.32 | -0.31 |
| AC018648.1 | -0.31 | -0.37 |
| AIM2 | -0.31 | -0.39 |
| ZNF775 | -0.31 | -0.33 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | IEA | glutamate (GO term level: 7) | - |
| GO:0008233 | peptidase activity | IEA | - | |
| GO:0042277 | peptide binding | ISS | 8940009 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002001 | renin secretion into blood stream | IEP | 8901832 | |
| GO:0001822 | kidney development | IMP | 18519639 | |
| GO:0001822 | kidney development | ISS | 8940009 | |
| GO:0006465 | signal peptide processing | IDA | 16912035 | |
| GO:0007267 | cell-cell signaling | TAS | 8901832 | |
| GO:0009952 | anterior/posterior pattern formation | IMP | 18519639 | |
| GO:0007507 | heart development | ISS | 8940009 | |
| GO:0007566 | embryo implantation | ISS | 8940009 | |
| GO:0051605 | protein maturation via proteolysis | IDA | 8901832 | |
| GO:0042089 | cytokine biosynthetic process | ISS | 8940009 | |
| GO:0019067 | viral assembly, maturation, egress, and release | IEP | 8940009 | |
| GO:0019067 | viral assembly, maturation, egress, and release | ISS | 8940009 | |
| GO:0030323 | respiratory tube development | ISS | 8940009 | |
| GO:0032455 | nerve growth factor processing | EXP | 12787574 | |
| GO:0035108 | limb morphogenesis | ISS | 8940009 | |
| GO:0043043 | peptide biosynthetic process | IDA | 8901832 | |
| GO:0048566 | embryonic gut development | IMP | 18519639 | |
| GO:0048566 | embryonic gut development | ISS | 8940009 | |
| GO:0048706 | embryonic skeletal system development | IMP | 18519639 | |
| GO:0048706 | embryonic skeletal system development | ISS | 8940009 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | ISS | - | |
| GO:0005796 | Golgi lumen | EXP | 8615794 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | ISS | 8940009 | |
| GO:0030141 | secretory granule | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| NEWMAN ERCC6 TARGETS UP | 26 | 18 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| WATANABE ULCERATIVE COLITIS WITH CANCER UP | 18 | 10 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS UP | 48 | 32 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP | 54 | 38 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-139 | 26 | 32 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.