Gene Page: PRKCZ
Summary ?
| GeneID | 5590 |
| Symbol | PRKCZ |
| Synonyms | PKC-ZETA|PKC2 |
| Description | protein kinase C zeta |
| Reference | MIM:176982|HGNC:HGNC:9412|Ensembl:ENSG00000067606|HPRD:01504|Vega:OTTHUMG00000001238 |
| Gene type | protein-coding |
| Map location | 1p36.33-p36.2 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.994 |
| Fetal beta | 1.517 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 7 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.9141 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01974180 | 1 | 2084246 | PRKCZ | 2.66E-5 | 0.468 | 0.018 | DMG:Wockner_2014 |
| cg04347414 | 1 | 2084519 | PRKCZ | 7.42E-5 | 0.628 | 0.025 | DMG:Wockner_2014 |
| cg02393699 | 1 | 2106620 | PRKCZ | 1.142E-4 | 0.412 | 0.029 | DMG:Wockner_2014 |
| cg00300046 | 1 | 2084595 | PRKCZ | 1.387E-4 | 0.671 | 0.031 | DMG:Wockner_2014 |
| cg24445388 | 1 | 2084391 | PRKCZ | 1.412E-4 | 0.743 | 0.031 | DMG:Wockner_2014 |
| cg17426766 | 1 | 2046864 | PRKCZ | 4.131E-4 | 0.365 | 0.044 | DMG:Wockner_2014 |
| cg18269141 | 1 | 2063966 | PRKCZ | 5.803E-4 | 0.353 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
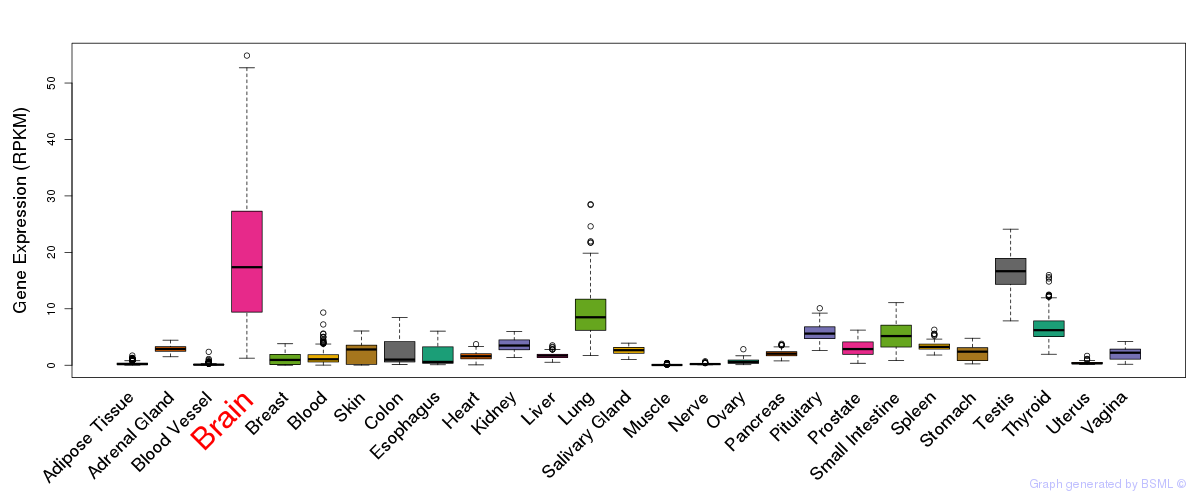
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004700 | atypical protein kinase C activity | TAS | 8224878 | |
| GO:0005515 | protein binding | IPI | 11755531 |12893243 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0019992 | diacylglycerol binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000226 | microtubule cytoskeleton organization | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | TAS | 10477520 | |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0006916 | anti-apoptosis | TAS | 10770953 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | IEA | Brain (GO term level: 10) | - |
| GO:0005624 | membrane fraction | TAS | 7925449 | |
| GO:0005737 | cytoplasm | TAS | 7925449 | |
| GO:0005768 | endosome | IEA | - | |
| GO:0005938 | cell cortex | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 7925449 | |
| GO:0045179 | apical cortex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADAP1 | CENTA1 | GCS1L | p42IP4 | ArfGAP with dual PH domains 1 | - | HPRD,BioGRID | 12893243 |
| ADCY5 | AC5 | adenylate cyclase 5 | - | HPRD | 8206971 |
| AFAP1 | AFAP | AFAP-110 | FLJ56849 | actin filament associated protein 1 | - | HPRD | 12134071 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 10085094 |
| AKT3 | DKFZp434N0250 | PKB-GAMMA | PKBG | PRKBG | RAC-PK-gamma | RAC-gamma | STK-2 | v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma) | - | HPRD,BioGRID | 12162751 |
| C1QBP | GC1QBP | HABP1 | SF2p32 | gC1Q-R | gC1qR | p32 | complement component 1, q subcomponent binding protein | - | HPRD,BioGRID | 10831594 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | - | HPRD | 10733591 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | - | HPRD,BioGRID | 10764587 |
| DAPK3 | FLJ36473 | ZIP | ZIPK | death-associated protein kinase 3 | Reconstituted Complex | BioGRID | 12242277 |
| ECT2 | FLJ10461 | MGC138291 | epithelial cell transforming sequence 2 oncogene | Affinity Capture-Western | BioGRID | 15254234 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD,BioGRID | 11739185 |
| FEZ1 | - | fasciculation and elongation protein zeta 1 (zygin I) | - | HPRD,BioGRID | 9971736 |
| FEZ2 | HUM3CL | MGC117372 | fasciculation and elongation protein zeta 2 (zygin II) | - | HPRD,BioGRID | 14697253 |
| GRB14 | - | growth factor receptor-bound protein 14 | - | HPRD,BioGRID | 12242277 |
| HABP4 | IHABP4 | Ki-1/57 | MGC825 | SERBP1L | hyaluronan binding protein 4 | - | HPRD | 14699138 |
| HNRNPA1 | HNRPA1 | MGC102835 | heterogeneous nuclear ribonucleoprotein A1 | PKC-zeta phosphorylates and interacts with hnRNPA1. This interaction was modeled on a demonstrated interaction between PKC-zeta and hnRNPA1, both from unspecified species. | BIND | 10383403 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | - | HPRD,BioGRID | 11937546 |
| IRS4 | IRS-4 | PY160 | insulin receptor substrate 4 | - | HPRD | 12774026 |
| KRT10 | CK10 | K10 | KPP | keratin 10 | - | HPRD,BioGRID | 11585925 |
| MAP2K5 | HsT17454 | MAPKK5 | MEK5 | PRKMK5 | mitogen-activated protein kinase kinase 5 | - | HPRD,BioGRID | 11158308 |
| MBP | MGC99675 | myelin basic protein | PKC-zeta phosphorylates and interacts with MBP. This interaction was modeled on a demonstrated interaction between PKC-zeta and MBP, both from unspecified species. | BIND | 10383403 |
| NCL | C23 | FLJ45706 | nucleolin | - | HPRD,BioGRID | 9388266 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD,BioGRID | 12021260 |
| PARD3 | ASIP | Baz | Bazooka | FLJ21015 | PAR3 | PAR3alpha | PARD3A | SE2-5L16 | SE2-5LT1 | SE2-5T2 | par-3 partitioning defective 3 homolog (C. elegans) | Affinity Capture-Western | BioGRID | 12459187 |
| PARD6A | PAR-6A | PAR6 | PAR6C | PAR6alpha | TAX40 | TIP-40 | par-6 partitioning defective 6 homolog alpha (C. elegans) | - | HPRD,BioGRID | 11260256 |
| PARD6G | FLJ45701 | PAR-6G | PAR6gamma | par-6 partitioning defective 6 homolog gamma (C. elegans) | - | HPRD,BioGRID | 11260256 |
| PAWR | PAR4 | Par-4 | PRKC, apoptosis, WT1, regulator | Par-4 interacts with PKC-zeta. This interaction was modelled on a demonstrated interaction between human Par-4 and rat PKC-zeta. | BIND | 8797824 |
| PAWR | PAR4 | Par-4 | PRKC, apoptosis, WT1, regulator | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8797824 |
| PDLIM7 | LMP1 | PDZ and LIM domain 7 (enigma) | - | HPRD | 8940095 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | PRKCZ (PKCZ) interacts with PDPK1 (PDK1). This interaction was modelled on a demonstrated interaction between human PRKCZ and rat PDPK1. | BIND | 11781095 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | An unspecified isoform of PDPK1 (PDK1) phosphorylates PRKCZ (PKC-zeta). This interaction was modeled on a demonstrated interaction between PDPK1 from an unspecified species and PRKCZ from an unspecified species. | BIND | 12223477 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | - | HPRD,BioGRID | 9748166 |10764742 |11781095 |
| PPP1R14A | CPI-17 | CPI17 | PPP1INL | protein phosphatase 1, regulatory (inhibitor) subunit 14A | - | HPRD | 15003508 |
| PPP3CA | CALN | CALNA | CALNA1 | CCN1 | CNA1 | PPP2B | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform | - | HPRD | 12021260 |
| PRKCSH | AGE-R2 | G19P1 | PCLD | PLD1 | protein kinase C substrate 80K-H | PRKCZ (PKCZ) interacts with PRKCSH (80K-H). | BIND | 15707389 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | PKC-zeta autophosphorylates. | BIND | 15665819 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 10620507 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 11684013 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Affinity Capture-Western | BioGRID | 16931574 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | zetaPKC interacts with p62. This interaction was modelled on a demonstrated interaction between rat zetaPKC and human p62. | BIND | 12813044 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | - | HPRD | 9566925 |10356400 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | Affinity Capture-Western | BioGRID | 10747026 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 10527887 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | Biochemical Activity | BioGRID | 10212259 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 10747026 |
| WWC1 | FLJ10865 | FLJ23369 | KIAA0869 | KIBRA | WW and C2 domain containing 1 | - | HPRD,BioGRID | 15081397 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD,BioGRID | 10620507 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Affinity Capture-Western | BioGRID | 10620507 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | Affinity Capture-Western | BioGRID | 10620507 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 10620507 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Biochemical Activity Reconstituted Complex | BioGRID | 10620507 |12893243 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTDINS PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID INSULIN PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID IL1 PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID IL2 PI3K PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID NEPHRIN NEPH1 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID RAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | 16 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB DN | 21 | 16 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD DN | 36 | 27 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| CUI GLUCOSE DEPRIVATION | 60 | 44 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K27ME3 | 79 | 59 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-338 | 203 | 209 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.