Gene Page: PRNP
Summary ?
| GeneID | 5621 |
| Symbol | PRNP |
| Synonyms | ASCR|AltPrP|CD230|CJD|GSS|KURU|PRIP|PrP|PrP27-30|PrP33-35C|PrPc|p27-30 |
| Description | prion protein |
| Reference | MIM:176640|HGNC:HGNC:9449|Ensembl:ENSG00000171867|HPRD:01453|Vega:OTTHUMG00000031786 |
| Gene type | protein-coding |
| Map location | 20p13 |
| Pascal p-value | 0.508 |
| Sherlock p-value | 0.878 |
| Fetal beta | -1.793 |
| DMG | 3 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 16.3147 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15993674 | 20 | 4667405 | PRNP | -0.02 | 0.98 | DMG:Nishioka_2013 | |
| cg15993674 | 20 | 4667405 | PRNP | 4.49E-10 | -0.014 | 8.02E-7 | DMG:Jaffe_2016 |
| cg09614401 | 20 | 4630022-4630071 | PRNP | 0.003 | 5.522 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
General gene expression (GTEx)
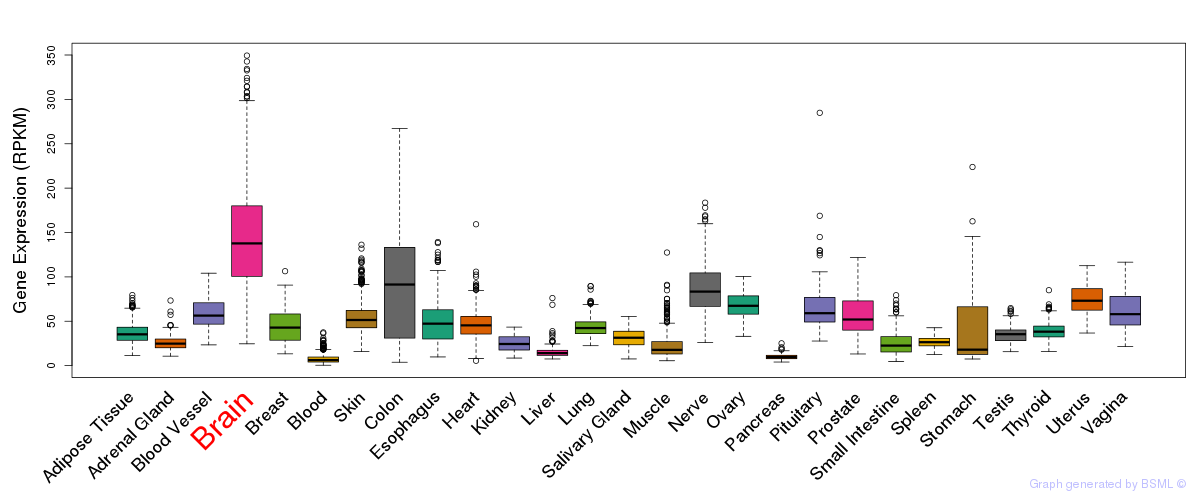
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TF | 0.97 | 0.95 |
| FA2H | 0.97 | 0.94 |
| GJB1 | 0.96 | 0.95 |
| SLC5A11 | 0.96 | 0.92 |
| MAG | 0.95 | 0.94 |
| SLC31A2 | 0.95 | 0.77 |
| CNP | 0.94 | 0.88 |
| PPAP2C | 0.94 | 0.90 |
| HSPA2 | 0.94 | 0.89 |
| DBNDD2 | 0.94 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NKIRAS2 | -0.55 | -0.72 |
| TUBB2B | -0.53 | -0.79 |
| HN1 | -0.53 | -0.75 |
| CRMP1 | -0.52 | -0.71 |
| KIAA1949 | -0.52 | -0.75 |
| NR2C2AP | -0.52 | -0.77 |
| TRNAU1AP | -0.52 | -0.73 |
| RARS2 | -0.52 | -0.74 |
| HMGB3 | -0.51 | -0.76 |
| IFT52 | -0.51 | -0.74 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005507 | copper ion binding | IEA | - | |
| GO:0005507 | copper ion binding | TAS | 16294306 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0008017 | microtubule binding | IDA | 16004966 | |
| GO:0042802 | identical protein binding | IPI | 16286452 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008152 | metabolic process | TAS | 3755672 | |
| GO:0006979 | response to oxidative stress | IEA | - | |
| GO:0006979 | response to oxidative stress | ISS | - | |
| GO:0006878 | cellular copper ion homeostasis | NAS | 16004966 | |
| GO:0051260 | protein homooligomerization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005794 | Golgi apparatus | ISS | - | |
| GO:0005737 | cytoplasm | TAS | 16004966 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0005783 | endoplasmic reticulum | ISS | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005886 | plasma membrane | ISS | - | |
| GO:0031225 | anchored to membrane | IEA | - | |
| GO:0019898 | extrinsic to membrane | TAS | 16004966 | |
| GO:0045121 | membrane raft | IEA | - | |
| GO:0045121 | membrane raft | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | - | HPRD | 15146195 |
| BAT3 | BAG-6 | BAG6 | D6S52E | G3 | HLA-B associated transcript 3 | Two-hybrid | BioGRID | 16169070 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD | 10988071 |
| CLSTN1 | ALC-ALPHA | CSTN1 | FLJ32258 | KIAA0911 | PIK3CD | XB31alpha | alcalpha1 | alcalpha2 | calsyntenin 1 | - | HPRD | 15146195 |
| CLU | AAG4 | APOJ | CLI | KUB1 | MGC24903 | SGP-2 | SGP2 | SP-40 | TRPM-2 | TRPM2 | clusterin | - | HPRD | 15146195 |
| CNTN1 | F3 | GP135 | contactin 1 | - | HPRD | 15146195 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | PrPc interacts with CSNK2A1 (CK2 alpha). This interaction was modeled on a demonstrated interaction between bovine PrPc and human CSNK2A1 (CK2 alpha). | BIND | 11062072 |
| CSNK2A2 | CK2A2 | CSNK2A1 | FLJ43934 | casein kinase 2, alpha prime polypeptide | PrPc interacts with CSNK2A2 (CK2 alpha prime). This interaction was modeled on a demonstrated interaction between bovine PrPc and human CSNK2A2 (CK2 alpha prime). | BIND | 11062072 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | PrPc interacts with CSNK2B (CK2 beta) albeit weakly. This interaction was modeled on a demonstrated interaction between bovine PrPc and human CSNK2B (CK2 beta). | BIND | 11062072 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 15146195 |
| DPP6 | DPPX | MGC46605 | dipeptidyl-peptidase 6 | - | HPRD | 15146195 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 11571277 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | PrPc interacts with HSPA5 (BiP). | BIND | 10970892 |
| HSPD1 | CPN60 | GROEL | HSP60 | HSP65 | HuCHA60 | SPG13 | heat shock 60kDa protein 1 (chaperonin) | PrPc interacts with HSPD1 (Hsp60). This interaction was modeled on a demonstrated interaction between hamster PrPc and human HSPD1 (Hsp60). | BIND | 8676499 |
| L1CAM | CAML1 | CD171 | HSAS | HSAS1 | MASA | MIC5 | N-CAML1 | S10 | SPG1 | L1 cell adhesion molecule | - | HPRD | 15146195 |
| MAG | GMA | S-MAG | SIGLEC-4A | SIGLEC4A | myelin associated glycoprotein | - | HPRD | 15146195 |
| MOG | MGC26137 | MOGIG-2 | myelin oligodendrocyte glycoprotein | - | HPRD | 15146195 |
| PLG | DKFZp779M0222 | plasminogen | PrPSc interacts with PLG. | BIND | 11438139 |
| PRNPIP | FLJ22943 | MGC2683 | PINT1 | prion protein interacting protein | - | HPRD | 11571277 |
| STIP1 | HOP | IEF-SSP-3521 | P60 | STI1 | STI1L | stress-induced-phosphoprotein 1 | - | HPRD,BioGRID | 12093732 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| PID GLYPICAN 1PATHWAY | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM1 INTERACTIONS | 39 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLACK UP | 35 | 22 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL DN | 42 | 25 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE DN | 47 | 34 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 LASER DN | 50 | 38 | All SZGR 2.0 genes in this pathway |
| HUI MAPK14 TARGETS UP | 21 | 11 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 6 | 75 | 48 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |