Gene Page: PTN
Summary ?
| GeneID | 5764 |
| Symbol | PTN |
| Synonyms | HARP|HBGF8|HBNF|NEGF1 |
| Description | pleiotrophin |
| Reference | MIM:162095|HGNC:HGNC:9630|Ensembl:ENSG00000105894|HPRD:01199|Vega:OTTHUMG00000155709 |
| Gene type | protein-coding |
| Map location | 7q33 |
| Pascal p-value | 1.221E-6 |
| Sherlock p-value | 0.766 |
| Fetal beta | 0.663 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.7318 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02701024 | 7 | 136989898 | PTN | 1.21E-5 | -0.496 | 0.014 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17014160 | chr1 | 206855799 | PTN | 5764 | 0.16 | trans | ||
| rs1394545 | chr4 | 136709578 | PTN | 5764 | 0.11 | trans | ||
| rs11730105 | chr4 | 136710424 | PTN | 5764 | 0.15 | trans | ||
| rs11953760 | chr5 | 9842943 | PTN | 5764 | 0.1 | trans |
Section II. Transcriptome annotation
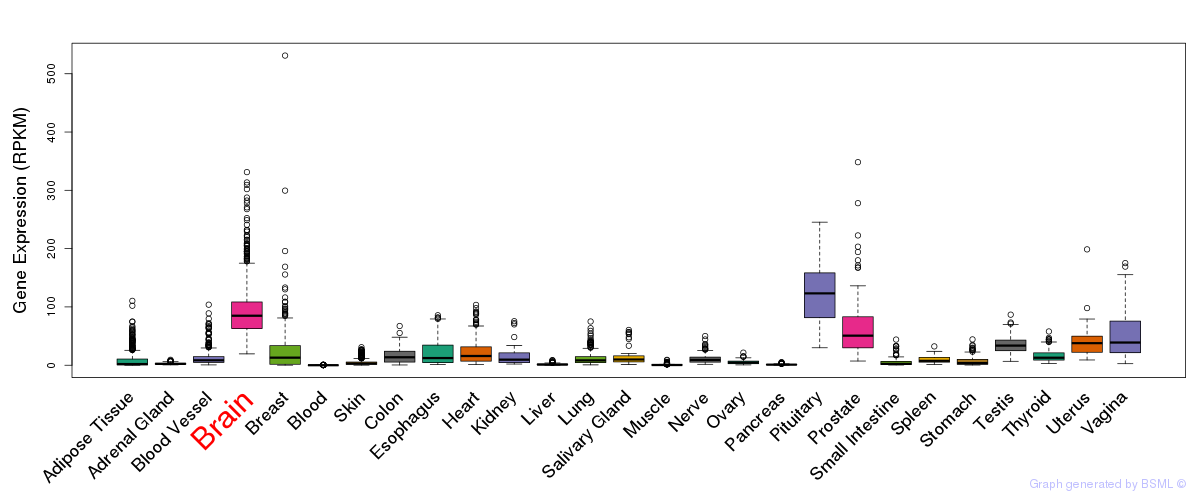
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LGALS8 | 0.86 | 0.81 |
| AC079061.1 | 0.86 | 0.81 |
| PTPN13 | 0.85 | 0.85 |
| DOCK9 | 0.85 | 0.88 |
| RNF19A | 0.82 | 0.81 |
| RP11-791G16.2 | 0.82 | 0.82 |
| MEF2C | 0.81 | 0.77 |
| ACTN2 | 0.80 | 0.77 |
| KLHL4 | 0.80 | 0.84 |
| RBM24 | 0.80 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DCXR | -0.52 | -0.56 |
| METRN | -0.52 | -0.62 |
| ENHO | -0.52 | -0.64 |
| HSD17B14 | -0.52 | -0.56 |
| REEP6 | -0.51 | -0.46 |
| AF347015.31 | -0.51 | -0.54 |
| ACSF2 | -0.51 | -0.56 |
| MT-CO2 | -0.50 | -0.54 |
| FXYD1 | -0.50 | -0.52 |
| TLCD1 | -0.49 | -0.55 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004864 | protein phosphatase inhibitor activity | TAS | 10706604 | |
| GO:0008201 | heparin binding | IEA | - | |
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 2270483 |
| GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway | TAS | 10706604 | |
| GO:0008284 | positive regulation of cell proliferation | TAS | 2270483 | |
| GO:0007612 | learning | IEA | - | |
| GO:0030282 | bone mineralization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | TAS | 10706604 | |
| GO:0005783 | endoplasmic reticulum | IDA | 11256614 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 9739080 |
| ALK | CD246 | Ki-1 | TFG/ALK | anaplastic lymphoma receptor tyrosine kinase | - | HPRD,BioGRID | 11278720 |
| APOBEC3G | ARP9 | CEM15 | FLJ12740 | MDS019 | bK150C2.7 | dJ494G10.1 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G | Two-hybrid | BioGRID | 16169070 |
| ARIH2 | ARI2 | FLJ10938 | FLJ33921 | TRIAD1 | ariadne homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| ARL15 | ARFRP2 | FLJ20051 | ADP-ribosylation factor-like 15 | Two-hybrid | BioGRID | 16169070 |
| ATP5C1 | ATP5C | ATP5CL1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | Two-hybrid | BioGRID | 16169070 |
| ATRX | ATR2 | MGC2094 | MRXHF1 | RAD54 | RAD54L | SFM1 | SHS | XH2 | XNP | ZNF-HX | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| BCCIP | TOK-1 | TOK1 | BRCA2 and CDKN1A interacting protein | Two-hybrid | BioGRID | 16169070 |
| BLOC1S1 | BLOS1 | FLJ39337 | FLJ97089 | GCN5L1 | MGC87455 | MICoA | RT14 | biogenesis of lysosomal organelles complex-1, subunit 1 | Two-hybrid | BioGRID | 16169070 |
| BMI1 | MGC12685 | PCGF4 | RNF51 | BMI1 polycomb ring finger oncogene | Two-hybrid | BioGRID | 16169070 |
| BTBD2 | - | BTB (POZ) domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| C8orf30A | FLJ40907 | chromosome 8 open reading frame 30A | Two-hybrid | BioGRID | 16169070 |
| C9orf82 | FLJ13657 | RP11-337A23.1 | chromosome 9 open reading frame 82 | Two-hybrid | BioGRID | 16169070 |
| CACNB4 | CAB4 | CACNLB4 | EA5 | EJM | calcium channel, voltage-dependent, beta 4 subunit | Two-hybrid | BioGRID | 16169070 |
| CAPZB | CAPB | CAPPB | CAPZ | MGC104401 | MGC129749 | MGC129750 | capping protein (actin filament) muscle Z-line, beta | Two-hybrid | BioGRID | 16169070 |
| CCDC53 | CGI-116 | coiled-coil domain containing 53 | Two-hybrid | BioGRID | 16169070 |
| CDH10 | - | cadherin 10, type 2 (T2-cadherin) | Two-hybrid | BioGRID | 16169070 |
| CDK5RAP2 | C48 | Cep215 | DKFZp686B1070 | DKFZp686D1070 | KIAA1633 | MCPH3 | CDK5 regulatory subunit associated protein 2 | Two-hybrid | BioGRID | 16169070 |
| CGA | CG-ALPHA | FSHA | GPHA1 | GPHa | HCG | LHA | TSHA | glycoprotein hormones, alpha polypeptide | Two-hybrid | BioGRID | 16169070 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| COX6C | - | cytochrome c oxidase subunit VIc | Two-hybrid | BioGRID | 16169070 |
| CRCT1 | C1orf42 | NICE-1 | cysteine-rich C-terminal 1 | Two-hybrid | BioGRID | 16169070 |
| CRELD1 | AVSD2 | CIRRIN | DKFZp566D213 | cysteine-rich with EGF-like domains 1 | Two-hybrid | BioGRID | 16169070 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 10706604 |
| DFNA5 | ICERE-1 | deafness, autosomal dominant 5 | Two-hybrid | BioGRID | 16169070 |
| DNAJB11 | ABBP-2 | ABBP2 | DJ9 | EDJ | ERdj3 | ERj3 | HEDJ | PRO1080 | UNQ537 | hDj9 | DnaJ (Hsp40) homolog, subfamily B, member 11 | Two-hybrid | BioGRID | 16169070 |
| DUSP12 | DUSP1 | YVH1 | dual specificity phosphatase 12 | Two-hybrid | BioGRID | 16169070 |
| EIF3F | EIF3S5 | eIF3-p47 | eukaryotic translation initiation factor 3, subunit F | Two-hybrid | BioGRID | 16169070 |
| FAM176A | FLJ13391 | TMEM166 | family with sequence similarity 176, member A | Two-hybrid | BioGRID | 16169070 |
| FTL | MGC71996 | ferritin, light polypeptide | Two-hybrid | BioGRID | 16169070 |
| GIPC2 | FLJ20075 | SEMCAP-2 | SEMCAP2 | GIPC PDZ domain containing family, member 2 | Two-hybrid | BioGRID | 16169070 |
| GLYAT | ACGNAT | CAT | GAT | glycine-N-acyltransferase | Two-hybrid | BioGRID | 16169070 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | Two-hybrid | BioGRID | 16169070 |
| GPD1 | FLJ26652 | glycerol-3-phosphate dehydrogenase 1 (soluble) | Two-hybrid | BioGRID | 16169070 |
| GSTP1 | DFN7 | FAEES3 | GST3 | PI | glutathione S-transferase pi 1 | Two-hybrid | BioGRID | 16169070 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Two-hybrid | BioGRID | 16169070 |
| HSPBAP1 | FLJ22623 | FLJ39386 | PASS1 | HSPB (heat shock 27kDa) associated protein 1 | Two-hybrid | BioGRID | 16169070 |
| ITSN2 | KIAA1256 | PRO2015 | SH3D1B | SH3P18 | SWA | SWAP | intersectin 2 | Two-hybrid | BioGRID | 16169070 |
| LAMA4 | DKFZp686D23145 | LAMA3 | LAMA4*-1 | laminin, alpha 4 | Two-hybrid | BioGRID | 16169070 |
| LUC7L2 | CGI-59 | CGI-74 | FLJ10657 | LUC7B2 | LUC7-like 2 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | Two-hybrid | BioGRID | 16169070 |
| MKKS | BBS6 | HMCS | KMS | MKS | McKusick-Kaufman syndrome | Two-hybrid | BioGRID | 16169070 |
| MLLT3 | AF9 | FLJ2035 | YEATS3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | Two-hybrid | BioGRID | 16169070 |
| MYCBP | AMY-1 | c-myc binding protein | Two-hybrid | BioGRID | 16169070 |
| NAP1L5 | DRLM | nucleosome assembly protein 1-like 5 | Two-hybrid | BioGRID | 16169070 |
| NCAN | CSPG3 | FLJ44681 | neurocan | - | HPRD | 9507007 |
| P4HB | DSI | ERBA2L | GIT | PDI | PDIA1 | PHDB | PO4DB | PO4HB | PROHB | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | Two-hybrid | BioGRID | 16169070 |
| PLXNB2 | KIAA0315 | MM1 | Nbla00445 | PLEXB2 | dJ402G11.3 | plexin B2 | Two-hybrid | BioGRID | 16169070 |
| PMF1 | - | polyamine-modulated factor 1 | Two-hybrid | BioGRID | 16169070 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | Two-hybrid | BioGRID | 16169070 |
| PSMB10 | LMP10 | MECL1 | MGC1665 | beta2i | proteasome (prosome, macropain) subunit, beta type, 10 | Two-hybrid | BioGRID | 16169070 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| PSMD2 | MGC14274 | P97 | Rpn1 | S2 | TRAP2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | Two-hybrid | BioGRID | 16169070 |
| PTPRB | DKFZp686E2262 | DKFZp686H15164 | FLJ44133 | HPTP-BETA | HPTPB | MGC142023 | MGC59935 | PTPB | R-PTP-BETA | VEPTP | protein tyrosine phosphatase, receptor type, B | - | HPRD | 10706604 |
| PTPRS | PTPSIGMA | protein tyrosine phosphatase, receptor type, S | Two-hybrid | BioGRID | 16169070 |
| PTPRZ1 | HPTPZ | HPTPzeta | PTP-ZETA | PTP18 | PTPRZ | PTPZ | RPTPB | RPTPbeta | phosphacan | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | - | HPRD,BioGRID | 10706604 |
| RBM23 | CAPERbeta | FLJ10482 | MGC4458 | PP239 | RNPC4 | RNA binding motif protein 23 | Two-hybrid | BioGRID | 16169070 |
| RIT1 | MGC125864 | MGC125865 | RIBB | RIT | ROC1 | Ras-like without CAAX 1 | Two-hybrid | BioGRID | 16169070 |
| RPLP1 | FLJ27448 | MGC5215 | P1 | RPP1 | ribosomal protein, large, P1 | Two-hybrid | BioGRID | 16169070 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| SDC1 | CD138 | SDC | SYND1 | syndecan | syndecan 1 | - | HPRD | 7592855 |
| SDC1 | CD138 | SDC | SYND1 | syndecan | syndecan 1 | Reconstituted Complex | BioGRID | 8175719 |
| SDC3 | N-syndecan | SDCN | SYND3 | syndecan 3 | - | HPRD | 8175719 |
| SGSM2 | KIAA0397 | RUTBC1 | small G protein signaling modulator 2 | Two-hybrid | BioGRID | 16169070 |
| SGTA | SGT | alphaSGT | hSGT | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | Two-hybrid | BioGRID | 16169070 |
| SLC25A36 | DKFZp564C053 | FLJ10618 | solute carrier family 25, member 36 | Two-hybrid | BioGRID | 16169070 |
| SNCAIP | MGC39814 | SYPH1 | synuclein, alpha interacting protein | Two-hybrid | BioGRID | 16169070 |
| SSR1 | DKFZp781N23103 | FLJ14232 | FLJ22100 | FLJ23034 | FLJ78242 | FLJ93042 | TRAPA | signal sequence receptor, alpha | Two-hybrid | BioGRID | 16169070 |
| STC2 | STC-2 | STCRP | stanniocalcin 2 | Two-hybrid | BioGRID | 16169070 |
| SUPT7L | ART1 | KIAA0764 | MGC90306 | SPT7L | STAF65 | STAF65(gamma) | suppressor of Ty 7 (S. cerevisiae)-like | Two-hybrid | BioGRID | 16169070 |
| TAC3 | NKB | NKNB | PRO1155 | ZNEUROK1 | tachykinin 3 | Two-hybrid | BioGRID | 16169070 |
| TAF1D | JOSD3 | MGC5306 | TAF(I)41 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa | Two-hybrid | BioGRID | 16169070 |
| TBRG4 | CPR2 | FASTKD4 | KIAA0948 | transforming growth factor beta regulator 4 | Two-hybrid | BioGRID | 16169070 |
| TOMM20 | KIAA0016 | MAS20 | MGC117367 | MOM19 | TOM20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | Two-hybrid | BioGRID | 16169070 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | Two-hybrid | BioGRID | 16169070 |
| TXNDC9 | APACD | thioredoxin domain containing 9 | Two-hybrid | BioGRID | 16169070 |
| UBQLN4 | A1U | C1orf6 | CIP75 | UBIN | ubiquilin 4 | Two-hybrid | BioGRID | 16169070 |
| UTP3 | CRL1 | CRLZ1 | FLJ23256 | SAS10 | UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| WFDC2 | HE4 | MGC57529 | WAP5 | dJ461P17.6 | WAP four-disulfide core domain 2 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| PEPPER CHRONIC LYMPHOCYTIC LEUKEMIA UP | 33 | 28 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP | 97 | 60 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| PATTERSON DOCETAXEL RESISTANCE | 29 | 20 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD DN | 36 | 27 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS RESISTANT UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS REVERTED DN | 5 | 5 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS DN | 64 | 41 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| HE PTEN TARGETS DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 6HR UP | 10 | 8 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM | 100 | 51 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 1 DN | 63 | 39 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 419 | 425 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-384 | 644 | 651 | 1A,m8 | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-496 | 338 | 344 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.