Gene Page: PTPN11
Summary ?
| GeneID | 5781 |
| Symbol | PTPN11 |
| Synonyms | BPTP3|CFC|JMML|METCDS|NS1|PTP-1D|PTP2C|SH-PTP2|SH-PTP3|SHP2 |
| Description | protein tyrosine phosphatase, non-receptor type 11 |
| Reference | MIM:176876|HGNC:HGNC:9644|Ensembl:ENSG00000179295|HPRD:01470|Vega:OTTHUMG00000134334 |
| Gene type | protein-coding |
| Map location | 12q24 |
| Pascal p-value | 0.209 |
| Sherlock p-value | 7.809E-4 |
| Fetal beta | -0.506 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.9666 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
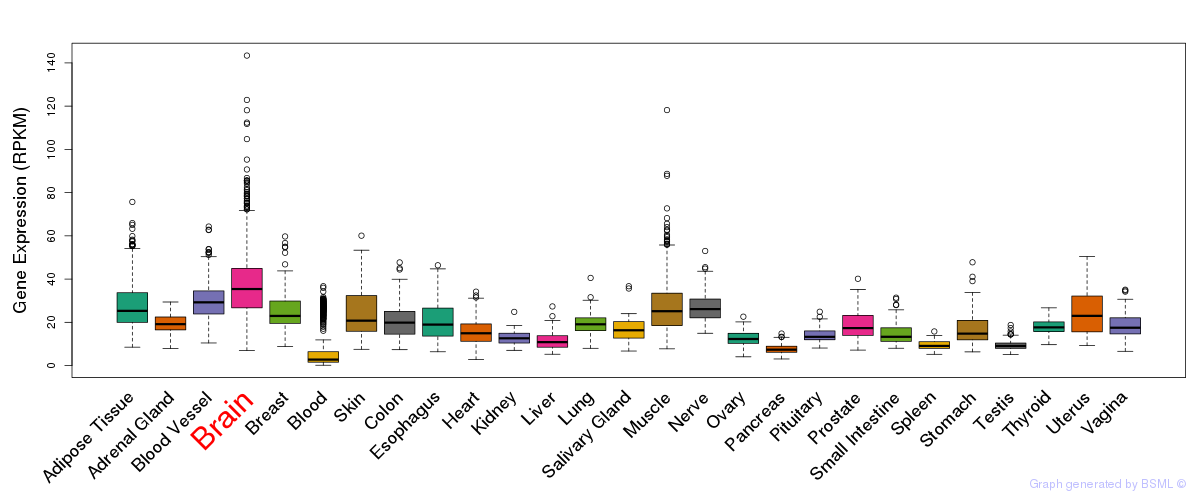
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| OTUD7A | 0.88 | 0.82 |
| TACC1 | 0.87 | 0.82 |
| ZMAT3 | 0.85 | 0.80 |
| MBNL1 | 0.84 | 0.80 |
| KIAA1671 | 0.83 | 0.79 |
| CHIC1 | 0.83 | 0.80 |
| RAPGEF5 | 0.83 | 0.73 |
| NR3C1 | 0.82 | 0.77 |
| FAM120A | 0.82 | 0.82 |
| MBNL2 | 0.82 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EXOSC8 | -0.47 | -0.53 |
| SNHG12 | -0.47 | -0.62 |
| RPL27 | -0.46 | -0.56 |
| C9orf46 | -0.46 | -0.54 |
| ST20 | -0.45 | -0.54 |
| RPL35 | -0.45 | -0.54 |
| PFDN5 | -0.44 | -0.52 |
| RPL31 | -0.44 | -0.55 |
| RPS13P2 | -0.44 | -0.54 |
| RPS23 | -0.44 | -0.55 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 9658397 |9792637 |10209036 | |
| GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity | EXP | 10734310 |14560030 |14665621 |15574420 | |
| GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity | TAS | 7681589 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0051428 | peptide hormone receptor binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007409 | axonogenesis | IEA | neuron, axon, neurite (GO term level: 12) | - |
| GO:0000077 | DNA damage checkpoint | IEA | - | |
| GO:0000187 | activation of MAPK activity | IEA | - | |
| GO:0009967 | positive regulation of signal transduction | IEA | - | |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| GO:0009755 | hormone-mediated signaling | IEA | - | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0048011 | nerve growth factor receptor signaling pathway | IEA | - | |
| GO:0006641 | triacylglycerol metabolic process | IEA | - | |
| GO:0006629 | lipid metabolic process | IEA | - | |
| GO:0042593 | glucose homeostasis | IEA | - | |
| GO:0042445 | hormone metabolic process | IEA | - | |
| GO:0035265 | organ growth | IEA | - | |
| GO:0040014 | regulation of multicellular organism growth | IEA | - | |
| GO:0046887 | positive regulation of hormone secretion | IEA | - | |
| GO:0046676 | negative regulation of insulin secretion | IEA | - | |
| GO:0046825 | regulation of protein export from nucleus | IEA | - | |
| GO:0048609 | reproductive process in a multicellular organism | IEA | - | |
| GO:0060125 | negative regulation of growth hormone secretion | IEA | - | |
| GO:0051463 | negative regulation of cortisol secretion | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9054388 |10734310 |14560030 |14665621 |15574420 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-Western | BioGRID | 9188452 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | Phosphorylated p210bcr-abl interacts with phosphorylated Syp. This interaction was modeled on a demonstrated interaction between human p210bcr-abl and mouse Syp. | BIND | 8195176 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | - | HPRD | 12115002 |
| BDKRB2 | B2R | BK-2 | BK2 | BKR2 | BRB2 | DKFZp686O088 | bradykinin receptor B2 | - | HPRD | 12177051 |
| BTLA | BTLA1 | CD272 | FLJ16065 | MGC129743 | B and T lymphocyte associated | - | HPRD,BioGRID | 12796776 |
| C6orf25 | G6b | MGC142279 | MGC142281 | NG31 | chromosome 6 open reading frame 25 | - | HPRD,BioGRID | 11544253 |
| CAT | MGC138422 | MGC138424 | catalase | Cat interacts with Shp-2. | BIND | 15556604 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 12176037 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | caveolin-1 interacts with SH-PTP2. | BIND | 12176037 |
| CD33 | FLJ00391 | SIGLEC-3 | SIGLEC3 | p67 | CD33 molecule | - | HPRD,BioGRID | 10206955 |
| CD84 | DKFZp781E2378 | LY9B | SLAMF5 | hCD84 | mCD84 | CD84 molecule | Affinity Capture-Western | BioGRID | 11689425 |
| CDH5 | 7B4 | CD144 | FLJ17376 | cadherin 5, type 2 (vascular endothelium) | - | HPRD,BioGRID | 10681592 |
| CEACAM1 | BGP | BGP1 | BGPI | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | - | HPRD,BioGRID | 9867848 |12108545 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD,BioGRID | 9344843 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | - | HPRD,BioGRID | 9162089 |
| CSF3R | CD114 | GCSFR | colony stimulating factor 3 receptor (granulocyte) | - | HPRD,BioGRID | 9824671 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | - | HPRD,BioGRID | 8638161 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 10681592 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 7673163 |
| EPHA2 | ECK | EPH receptor A2 | Affinity Capture-Western | BioGRID | 10655584 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD,BioGRID | 8639815 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | Affinity Capture-MS | BioGRID | 16729043 |
| ERBB4 | HER4 | MGC138404 | p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | Affinity Capture-MS | BioGRID | 16729043 |
| FCRL3 | FCRH3 | IFGP3 | IRTA3 | SPAP2 | Fc receptor-like 3 | Reconstituted Complex | BioGRID | 12051764 |
| FLT1 | FLT | VEGFR1 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | Flt-1 interacts with the amino terminal SH2 domain of SHP-2. | BIND | 9600074 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | - | HPRD | 9632781 |11447289 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | - | HPRD,BioGRID | 9632781 |
| FRS3 | FRS2B | FRS2beta | MGC17167 | SNT-2 | SNT2 | fibroblast growth factor receptor substrate 3 | - | HPRD | 11432792 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 10212213 |
| GAB1 | - | GRB2-associated binding protein 1 | SHP2 interacts with and dephosphorylates Gab1. | BIND | 12582165 |
| GAB1 | - | GRB2-associated binding protein 1 | - | HPRD,BioGRID | 11940581 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD | 10068651 |11287610 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | Affinity Capture-Western Two-hybrid | BioGRID | 10391903 |11334882 |11782427 |12135708 |
| GAB3 | - | GRB2-associated binding protein 3 | - | HPRD,BioGRID | 11739737 |
| GHR | GHBP | growth hormone receptor | - | HPRD,BioGRID | 10976913 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8041791 |8702859 |8995399 |9362449 |9632781 |9824671 |10080542 |10212213 |10747947 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 8041791 |8702859 |12531430 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Grb2 interacts with Syp via Syp SH2 domains. | BIND | 7523381 |8195176 |
| IFNAR1 | AVP | IFN-alpha-REC | IFNAR | IFNBR | IFRC | interferon (alpha, beta and omega) receptor 1 | - | HPRD | 9029147 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 7642582 |
| IL4R | CD124 | IL4RA | interleukin 4 receptor | - | HPRD,BioGRID | 11714803 |
| IL6ST | CD130 | CDw130 | GP130 | GP130-RAPS | IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | - | HPRD,BioGRID | 10946280 |
| IL6ST | CD130 | CDw130 | GP130 | GP130-RAPS | IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | - | HPRD | 9195977 |
| INPP5D | MGC104855 | MGC142140 | MGC142142 | SHIP | SHIP1 | SIP-145 | hp51CN | inositol polyphosphate-5-phosphatase, 145kDa | Affinity Capture-Western | BioGRID | 9110989 |
| INSR | CD220 | HHF5 | insulin receptor | - | HPRD,BioGRID | 7493946 |8135823 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 8505282 |9756938 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD,BioGRID | 8910607 |
| IRS4 | IRS-4 | PY160 | insulin receptor substrate 4 | - | HPRD | 12774026 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 8995399 |11036942 |12403768 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | Biochemical Activity in vivo Reconstituted Complex | BioGRID | 8639815 |8912646 |8995399 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD | 7559603 |8912646 |8995399 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 7523381 |9528781 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | Syp interacts with c-Kit via its SH2 domain. | BIND | 7523381 |
| KLRC1 | CD159A | MGC13374 | MGC59791 | NKG2 | NKG2A | killer cell lectin-like receptor subfamily C, member 1 | - | HPRD | 9485206 |
| LAIR1 | CD305 | LAIR-1 | leukocyte-associated immunoglobulin-like receptor 1 | - | HPRD,BioGRID | 9285412 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD | 10617656 |
| LEPR | CD295 | OBR | leptin receptor | - | HPRD | 11018044 |
| LIFR | CD118 | FLJ98106 | FLJ99923 | LIF-R | SJS2 | STWS | SWS | leukemia inhibitory factor receptor alpha | - | HPRD,BioGRID | 10800945 |
| LILRB4 | CD85K | HM18 | ILT3 | LILRB5 | LIR-5 | LIR5 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4 | - | HPRD | 9422771 |
| LILRB4 | CD85K | HM18 | ILT3 | LILRB5 | LIR-5 | LIR5 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4 | Protein-peptide | BioGRID | 9973385 |
| LY9 | CD229 | SLAMF3 | hly9 | mLY9 | lymphocyte antigen 9 | Affinity Capture-Western | BioGRID | 11689425 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | - | HPRD | 8662733 |
| MPZL1 | FLJ21047 | PZR | PZR1b | PZRa | PZRb | myelin protein zero-like 1 | - | HPRD,BioGRID | 9792637 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | - | HPRD | 8879209 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD | 10790433 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Reconstituted Complex Two-hybrid | BioGRID | 7691811 |11266449 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | PDGFR interacts with Syp. | BIND | 7688466 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Autophosphorylated PDGFR interacts with Syp. This interaction was modelled on a demonstrated interaction between human PDGFR and Syp from an unspecified species. | BIND | 8890167 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 9054388 |9774457 |10350061 |10801826 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Affinity Capture-Western | BioGRID | 9918857 |
| PILRA | FDF03 | paired immunoglobin-like type 2 receptor alpha | - | HPRD | 10660620 |
| PILRA | FDF03 | paired immunoglobin-like type 2 receptor alpha | Affinity Capture-Western | BioGRID | 10903717 |
| PLCG2 | - | phospholipase C, gamma 2 (phosphatidylinositol-specific) | Affinity Capture-Western | BioGRID | 12135708 |
| PRLR | hPRLrI | prolactin receptor | Affinity Capture-Western | BioGRID | 10991949 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 7589239 |8986614 |10082579 |10655584 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10880513 |
| PXN | FLJ16691 | paxillin | - | HPRD | 8986614 |10082579 |10655584 |
| ROS1 | MCF3 | ROS | c-ros-1 | c-ros oncogene 1 , receptor tyrosine kinase | Two-hybrid | BioGRID | 11266449 |
| SELE | CD62E | ELAM | ELAM1 | ESEL | LECAM2 | selectin E | - | HPRD,BioGRID | 11602579 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD | 10650943 |
| SIGLEC11 | - | sialic acid binding Ig-like lectin 11 | - | HPRD,BioGRID | 11986327 |
| SIGLEC12 | FLJ38600 | S2V | SIGLECL1 | SLG | Siglec-12 | Siglec-L1 | Siglec-XII | sialic acid binding Ig-like lectin 12 | - | HPRD | 11328818 |
| SIGLEC7 | AIRM1 | CD328 | CDw328 | D-siglec | QA79 | SIGLEC-7 | p75 | p75/AIRM1 | sialic acid binding Ig-like lectin 7 | - | HPRD | 10499918 |
| SIT1 | MGC125908 | MGC125909 | MGC125910 | RP11-331F9.5 | SIT | signaling threshold regulating transmembrane adaptor 1 | - | HPRD,BioGRID | 10209036 |11433379 |11491537 |
| SLAMF1 | CD150 | CDw150 | SLAM | signaling lymphocytic activation molecule family member 1 | - | HPRD,BioGRID | 11806999 |
| SLAMF1 | CD150 | CDw150 | SLAM | signaling lymphocytic activation molecule family member 1 | CD150 interacts with SHP-2. This interaction was modelled on a demonstrated interaction between human CD150 and monkey SHP-2. | BIND | 11477068 |
| SLAMF6 | KALI | KALIb | Ly108 | MGC104953 | NTB-A | NTBA | SF2000 | SLAM family member 6 | - | HPRD,BioGRID | 11489943 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12403768 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 9344843 |9632781 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 11594781 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 12060651 |
| STAT5B | STAT5 | signal transducer and activator of transcription 5B | Biochemical Activity Reconstituted Complex | BioGRID | 10617656 |
| SYNCRIP | GRY-RBP | HNRPQ1 | NSAP1 | RP1-3J17.2 | dJ3J17.2 | hnRNP-Q | pp68 | synaptotagmin binding, cytoplasmic RNA interacting protein | Two-hybrid | BioGRID | 9847309 |
| TEK | CD202B | TIE-2 | TIE2 | VMCM | VMCM1 | TEK tyrosine kinase, endothelial | - | HPRD | 10521483 |
| TIE1 | JTK14 | TIE | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | - | HPRD | 10949653 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD | 11786908 |
| TREML1 | GLTL1825 | MGC119173 | PRO3438 | TLT-1 | TLT1 | dJ238O23.3 | triggering receptor expressed on myeloid cells-like 1 | - | HPRD,BioGRID | 15128762 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 1752 | 1758 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-138 | 2277 | 2284 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-200bc/429 | 3739 | 3745 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-496 | 3824 | 3831 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.