Gene Page: RBBP5
Summary ?
| GeneID | 5929 |
| Symbol | RBBP5 |
| Synonyms | RBQ3|SWD1 |
| Description | retinoblastoma binding protein 5 |
| Reference | MIM:600697|HGNC:HGNC:9888|Ensembl:ENSG00000117222|HPRD:02826|Vega:OTTHUMG00000037104 |
| Gene type | protein-coding |
| Map location | 1q32 |
| Sherlock p-value | 0.093 |
| TADA p-value | 0.01 |
| Fetal beta | -0.021 |
| eGene | Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RBBP5 | chr1 | 205068937 | G | A | NM_001193272 NM_001193272 NM_001193273 NM_001193273 NM_005057 NM_005057 | . p.304H>Y . p.177H>Y . p.304H>Y | splice missense splice missense splice missense | Schizophrenia | DNM:Gulsuner_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2289256 | chr4 | 187089132 | RBBP5 | 5929 | 0.02 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
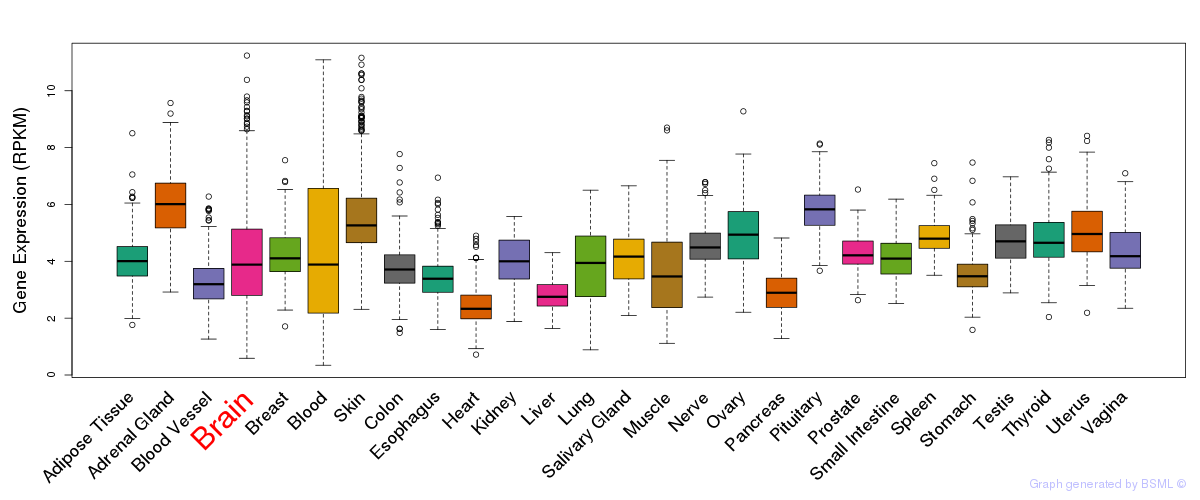
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRRG3 | 0.74 | 0.81 |
| L3MBTL2 | 0.74 | 0.79 |
| GGA3 | 0.74 | 0.80 |
| AP3B2 | 0.73 | 0.80 |
| AP2A2 | 0.73 | 0.79 |
| KCTD21 | 0.72 | 0.77 |
| USP11 | 0.72 | 0.79 |
| MAP3K12 | 0.72 | 0.80 |
| PISD | 0.72 | 0.77 |
| ZFYVE27 | 0.72 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.57 | -0.62 |
| AF347015.31 | -0.56 | -0.61 |
| AF347015.21 | -0.55 | -0.64 |
| HIGD1B | -0.55 | -0.62 |
| AC098691.2 | -0.54 | -0.66 |
| GNG11 | -0.53 | -0.64 |
| AF347015.33 | -0.53 | -0.57 |
| AC021016.1 | -0.53 | -0.59 |
| AF347015.8 | -0.53 | -0.60 |
| AF347015.2 | -0.53 | -0.57 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASCL2 | ASH2 | HASH2 | MASH2 | bHLHa45 | achaete-scute complex homolog 2 (Drosophila) | Affinity Capture-Western Co-purification | BioGRID | 12482968 |
| ASH2L | ASH2 | ASH2L1 | ASH2L2 | Bre2 | ash2 (absent, small, or homeotic)-like (Drosophila) | Reconstituted Complex | BioGRID | 15199122 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | Co-purification | BioGRID | 15199122 |
| HCFC2 | FLJ94012 | HCF-2 | HCF2 | host cell factor C2 | Co-purification | BioGRID | 15199122 |
| HIST2H3C | H3 | H3.2 | H3/M | H3F2 | H3FM | MGC9629 | histone cluster 2, H3c | Reconstituted Complex | BioGRID | 12482968 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | Co-purification | BioGRID | 15199122 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | RbBP5 interacts with MLL. | BIND | 15960975 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| MLL3 | DKFZp686C08112 | FLJ12625 | FLJ38309 | HALR | KIAA1506 | KMT2C | MGC119851 | MGC119852 | MGC119853 | myeloid/lymphoid or mixed-lineage leukemia 3 | Affinity Capture-Western Co-purification | BioGRID | 12482968 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 12482968 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 7558034 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | Co-purification | BioGRID | 12482968 |
| TUBB | M40 | MGC117247 | MGC16435 | OK/SW-cl.56 | TUBB1 | TUBB5 | tubulin, beta | Co-purification | BioGRID | 12482968 |
| WDR5 | BIG-3 | SWD3 | WD repeat domain 5 | Co-purification | BioGRID | 15199122 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| BARIS THYROID CANCER UP | 23 | 9 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| CLIMENT BREAST CANCER COPY NUMBER UP | 23 | 20 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |