| REACTOME SIGNALING BY GPCR
| 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING
| 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS
| 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Z SIGNALLING EVENTS
| 44 | 29 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN
| 169 | 118 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP
| 351 | 230 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN
| 145 | 91 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP
| 423 | 283 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP
| 74 | 45 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN
| 460 | 312 | All SZGR 2.0 genes in this pathway |
| NEWMAN ERCC6 TARGETS DN
| 39 | 24 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN
| 310 | 188 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN
| 367 | 220 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP
| 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP
| 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP
| 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP
| 164 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN
| 1375 | 806 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN
| 172 | 112 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP
| 185 | 119 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE
| 123 | 80 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN
| 712 | 443 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN
| 164 | 111 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP
| 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN
| 637 | 377 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP
| 256 | 159 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP
| 266 | 171 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN
| 200 | 112 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING DN
| 90 | 57 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN
| 289 | 166 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG DN
| 85 | 56 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS UP
| 52 | 34 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 UP
| 46 | 29 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN
| 162 | 102 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS DN
| 64 | 38 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN
| 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN
| 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN
| 371 | 218 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS UP
| 38 | 24 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN
| 101 | 70 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN
| 165 | 106 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP
| 566 | 371 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP
| 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP
| 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP
| 601 | 369 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN
| 108 | 68 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP
| 425 | 298 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS DN
| 68 | 48 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN
| 342 | 220 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP
| 395 | 249 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP
| 288 | 168 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY DN
| 38 | 27 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN
| 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN
| 88 | 68 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN
| 254 | 158 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP
| 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING
| 243 | 155 | All SZGR 2.0 genes in this pathway |
| PLASARI NFIC TARGETS BASAL UP
| 27 | 17 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN
| 106 | 77 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP
| 54 | 38 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP
| 221 | 120 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP
| 297 | 194 | All SZGR 2.0 genes in this pathway |
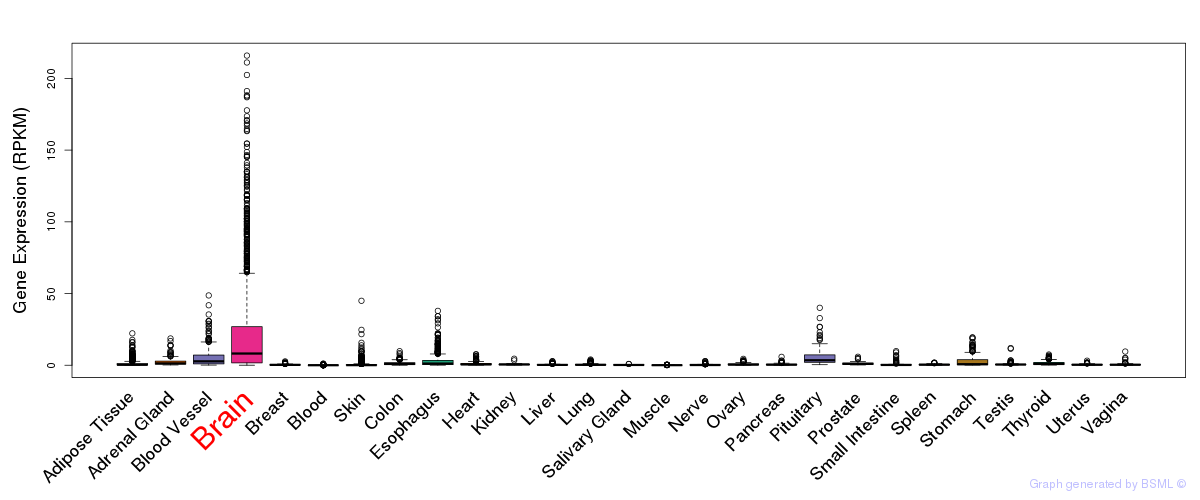
 eQTL annotation
eQTL annotation