Gene Page: RREB1
Summary ?
| GeneID | 6239 |
| Symbol | RREB1 |
| Synonyms | FINB|HNT|LZ321|RREB-1|Zep-1 |
| Description | ras responsive element binding protein 1 |
| Reference | MIM:602209|HGNC:HGNC:10449|Ensembl:ENSG00000124782|HPRD:03733|Vega:OTTHUMG00000014201 |
| Gene type | protein-coding |
| Map location | 6p25 |
| Pascal p-value | 0.035 |
| Sherlock p-value | 2.787E-4 |
| Fetal beta | -0.449 |
| DMG | 2 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08355568 | 6 | 7108248 | RREB1 | 2.968E-4 | -0.387 | 0.039 | DMG:Wockner_2014 |
| cg08651674 | 6 | 7108441 | RREB1 | -0.035 | 0.35 | DMG:Nishioka_2013 | |
| cg18533466 | 6 | 7107952 | RREB1 | -0.02 | 0.94 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
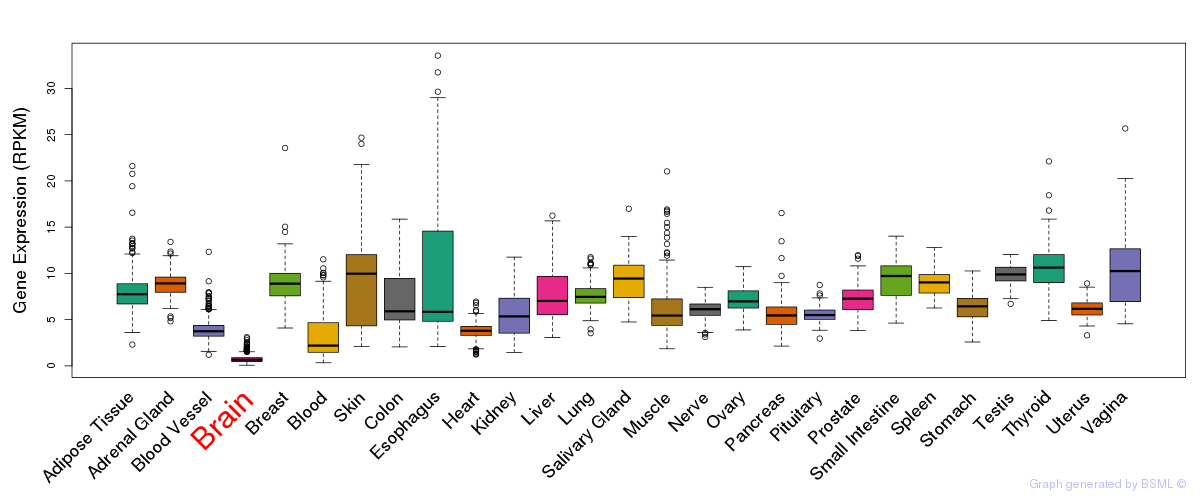
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAPGEF5 | 0.93 | 0.86 |
| LANCL1 | 0.90 | 0.89 |
| PRUNE2 | 0.90 | 0.82 |
| ENPP4 | 0.89 | 0.87 |
| ENDOD1 | 0.89 | 0.82 |
| FAM73A | 0.89 | 0.87 |
| ARFGEF2 | 0.89 | 0.88 |
| MFSD6 | 0.89 | 0.88 |
| CLIP4 | 0.89 | 0.82 |
| PCDH9 | 0.88 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.47 | -0.52 |
| AC006276.2 | -0.47 | -0.48 |
| RPL35 | -0.44 | -0.50 |
| RPL36 | -0.43 | -0.48 |
| TBC1D10A | -0.43 | -0.35 |
| ST20 | -0.42 | -0.49 |
| RPL12 | -0.42 | -0.42 |
| RPLP1 | -0.41 | -0.42 |
| C9orf46 | -0.41 | -0.41 |
| RPL31 | -0.40 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0016563 | transcription activator activity | TAS | 9305772 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | TAS | 9305772 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 8816445 | |
| GO:0007265 | Ras protein signal transduction | TAS | 8816445 | |
| GO:0007275 | multicellular organismal development | TAS | 8816445 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | TAS | 9305772 | |
| GO:0005737 | cytoplasm | TAS | 9305772 | |
| GO:0016607 | nuclear speck | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| MIDORIKAWA AMPLIFIED IN LIVER CANCER | 55 | 38 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE DN | 65 | 31 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR DN | 37 | 20 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 995 | 1001 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124.1 | 218 | 225 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 217 | 224 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-144 | 995 | 1001 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-145 | 457 | 464 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-155 | 335 | 341 | m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-23 | 472 | 478 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.