Gene Page: S100B
Summary ?
| GeneID | 6285 |
| Symbol | S100B |
| Synonyms | NEF|S100|S100-B|S100beta |
| Description | S100 calcium binding protein B |
| Reference | MIM:176990|HGNC:HGNC:10500|HPRD:01505| |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.033 |
| Sherlock p-value | 0.471 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0192 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1390969 | chr1 | 233690104 | S100B | 6285 | 0.15 | trans | ||
| rs357840 | chr4 | 60005852 | S100B | 6285 | 0.12 | trans | ||
| rs3884624 | chr6 | 96112071 | S100B | 6285 | 0 | trans | ||
| rs17695935 | chr7 | 140507936 | S100B | 6285 | 0.18 | trans | ||
| rs17070904 | chr18 | 60917055 | S100B | 6285 | 0.01 | trans |
Section II. Transcriptome annotation
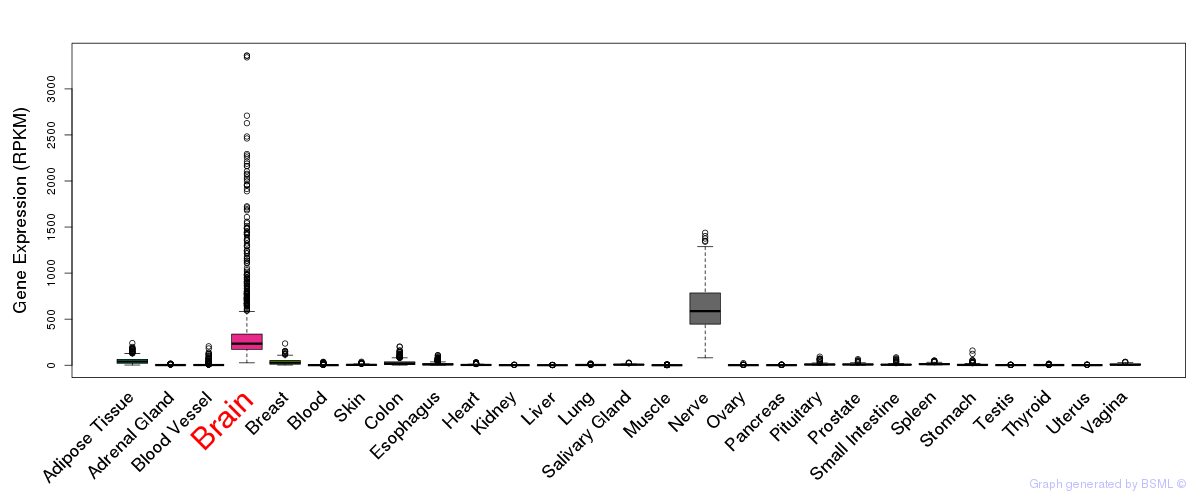
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C9orf142 | 0.79 | 0.72 |
| ZNF580 | 0.78 | 0.77 |
| SCAND1 | 0.78 | 0.75 |
| GAMT | 0.78 | 0.77 |
| AC008532.1 | 0.78 | 0.77 |
| C19orf20 | 0.78 | 0.78 |
| C4orf48 | 0.77 | 0.73 |
| YIF1B | 0.77 | 0.75 |
| RABAC1 | 0.76 | 0.72 |
| ZNHIT2 | 0.76 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CYB | -0.35 | -0.35 |
| AF347015.26 | -0.35 | -0.36 |
| MT-ATP8 | -0.35 | -0.40 |
| AF347015.8 | -0.35 | -0.34 |
| AF347015.27 | -0.34 | -0.34 |
| AF347015.15 | -0.33 | -0.33 |
| AF347015.33 | -0.33 | -0.33 |
| AF347015.2 | -0.33 | -0.33 |
| AF347015.31 | -0.33 | -0.32 |
| MT-CO2 | -0.32 | -0.31 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0048154 | S100 beta binding | IPI | astrocyte (GO term level: 4) | 9519411 |10913138 |
| GO:0005509 | calcium ion binding | NAS | 9519411 | |
| GO:0048306 | calcium-dependent protein binding | IDA | 10913138 | |
| GO:0008270 | zinc ion binding | ISS | - | |
| GO:0042803 | protein homodimerization activity | IDA | 10913138 | |
| GO:0042803 | protein homodimerization activity | IPI | 9519411 | |
| GO:0048156 | tau protein binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 8202493 |
| GO:0007409 | axonogenesis | TAS | neuron, axon, neurite (GO term level: 12) | 8202493 |
| GO:0048168 | regulation of neuronal synaptic plasticity | IEA | neuron, Synap (GO term level: 9) | - |
| GO:0008283 | cell proliferation | TAS | 8202493 | |
| GO:0007613 | memory | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IEA | axon, dendrite (GO term level: 4) | - |
| GO:0001726 | ruffle | IDA | 10913138 | |
| GO:0005634 | nucleus | IDA | 10913138 | |
| GO:0005737 | cytoplasm | IDA | 10913138 | |
| GO:0005737 | cytoplasm | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AGER | MGC22357 | RAGE | advanced glycosylation end product-specific receptor | - | HPRD | 10399917 |
| AHNAK | AHNAKRS | MGC5395 | AHNAK nucleoprotein | - | HPRD,BioGRID | 11312263 |
| ANXA6 | ANX6 | CBP68 | annexin A6 | - | HPRD,BioGRID | 9883272 |
| CACYBP | GIG5 | MGC87971 | PNAS-107 | RP1-102G20.6 | S100A6BP | SIP | calcyclin binding protein | - | HPRD | 12042313 |
| CAPZA1 | CAPPA1 | CAPZ | CAZ1 | capping protein (actin filament) muscle Z-line, alpha 1 | Reconstituted Complex | BioGRID | 12470955 |
| DES | CMD1I | CSM1 | CSM2 | FLJ12025 | FLJ39719 | FLJ41013 | FLJ41793 | desmin | - | HPRD | 10510252 |
| ECD | GCR2 | HSGT1 | ecdysoneless homolog (Drosophila) | - | HPRD | 12746458 |
| GFAP | FLJ45472 | glial fibrillary acidic protein | - | HPRD | 10510252 |
| IQGAP1 | HUMORFA01 | KIAA0051 | SAR1 | p195 | IQ motif containing GTPase activating protein 1 | - | HPRD,BioGRID | 12377780 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD | 11264299 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD,BioGRID | 2833519 |
| NDRG1 | CAP43 | CMT4D | DRG1 | GC4 | HMSNL | NDR1 | NMSL | PROXY1 | RIT42 | RTP | TARG1 | TDD5 | N-myc downstream regulated 1 | - | HPRD,BioGRID | 12493777 |
| PGM1 | - | phosphoglucomutase 1 | Affinity Capture-Western Far Western | BioGRID | 8894274 |
| S100A1 | S100 | S100-alpha | S100A | S100 calcium binding protein A1 | Affinity Capture-Western Two-hybrid | BioGRID | 9925766 |10913138 |16189514 |
| S100A1 | S100 | S100-alpha | S100A | S100 calcium binding protein A1 | S100A1 interacts with S100B. | BIND | 10913138 |
| S100A11 | MLN70 | S100C | S100 calcium binding protein A11 | - | HPRD,BioGRID | 10913138 |
| S100A11 | MLN70 | S100C | S100 calcium binding protein A11 | S100A11 interacts with S100B. | BIND | 10913138 |
| S100A6 | 2A9 | 5B10 | CABP | CACY | PRA | S100 calcium binding protein A6 | - | HPRD,BioGRID | 9925766 |10913138 |
| S100A6 | 2A9 | 5B10 | CABP | CACY | PRA | S100 calcium binding protein A6 | S100A6 interacts with S100B. | BIND | 10913138 |
| S100B | NEF | S100 | S100beta | S100 calcium binding protein B | Affinity Capture-Western Two-hybrid | BioGRID | 9925766 |10913138 |
| S100B | NEF | S100 | S100beta | S100 calcium binding protein B | S100B interacts with S100B. | BIND | 10913138 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western | BioGRID | 15178678 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 10876243 |11527429 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Affinity Capture-Western Reconstituted Complex | BioGRID | 10394361 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | 23 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED NFKB ACTIVATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN MONOCYTE DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION | 77 | 62 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP | 18 | 16 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS | 89 | 59 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |