Gene Page: SRPK1
Summary ?
| GeneID | 6732 |
| Symbol | SRPK1 |
| Synonyms | SFRSK1 |
| Description | SRSF protein kinase 1 |
| Reference | MIM:601939|HGNC:HGNC:11305|Ensembl:ENSG00000096063|HPRD:15992|Vega:OTTHUMG00000014583 |
| Gene type | protein-coding |
| Map location | 6p21.31 |
| Pascal p-value | 0.087 |
| Sherlock p-value | 0.509 |
| Fetal beta | 0.999 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09082921 | 6 | 35744226 | SRPK1 | 0.002 | -8 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9366911 | chr6 | 36589339 | SRPK1 | 6732 | 0.16 | cis |
Section II. Transcriptome annotation
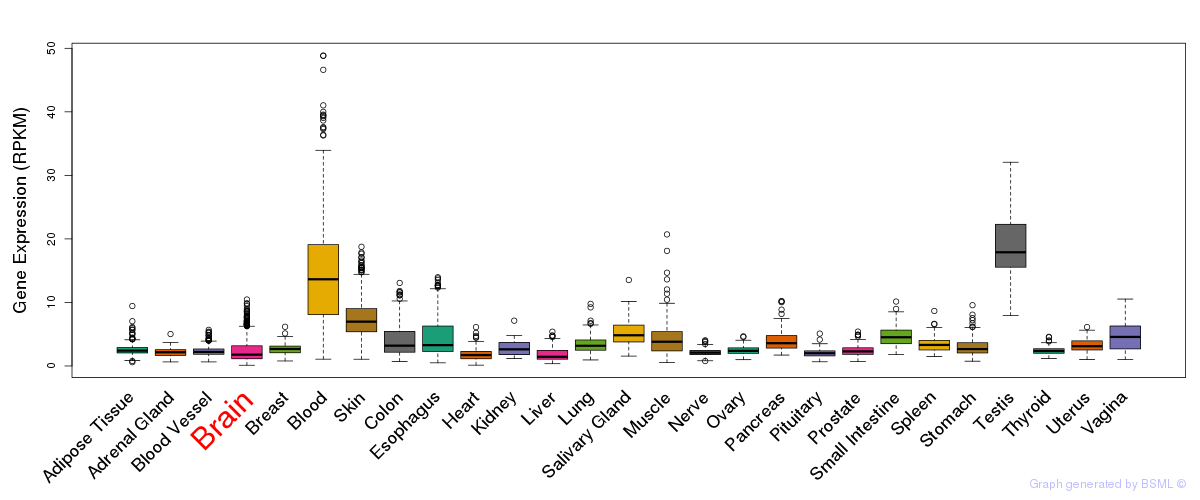
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HERC6 | 0.88 | 0.72 |
| MX1 | 0.80 | 0.55 |
| IFIT3 | 0.78 | 0.44 |
| IFIT1 | 0.77 | 0.51 |
| ATPAF1 | 0.75 | 0.73 |
| C1QB | 0.75 | 0.58 |
| CD59 | 0.75 | 0.65 |
| PPT1 | 0.75 | 0.81 |
| AC068989.2 | 0.75 | 0.61 |
| HSD17B12 | 0.74 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM88 | -0.41 | -0.47 |
| EGFL8 | -0.40 | -0.39 |
| RP9P | -0.38 | -0.53 |
| MYLK2 | -0.36 | -0.40 |
| AC005921.3 | -0.36 | -0.47 |
| AC011475.1 | -0.36 | -0.39 |
| SLC38A5 | -0.35 | -0.40 |
| AL022328.1 | -0.34 | -0.46 |
| RAMP2 | -0.34 | -0.44 |
| C1orf86 | -0.34 | -0.30 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IDA | 11509566 | |
| GO:0005515 | protein binding | IPI | 10196197 |11509566 |17353931 | |
| GO:0005524 | ATP binding | IDA | 11509566 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IDA | 11509566 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008380 | RNA splicing | TAS | 8208298 | |
| GO:0006468 | protein amino acid phosphorylation | IDA | 11509566 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007243 | protein kinase cascade | IDA | 11509566 | |
| GO:0007059 | chromosome segregation | IDA | 15034300 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0050684 | regulation of mRNA processing | IDA | 8208298 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 11509566 | |
| GO:0005737 | cytoplasm | IDA | 11509566 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CROP | LUC7A | OA48-18 | cisplatin resistance-associated overexpressed protein | Biochemical Activity | BioGRID | 12565863 |
| FERMT1 | C20orf42 | DTGCU2 | FLJ20116 | FLJ23423 | KIND1 | UNC112A | URP1 | fermitin family homolog 1 (Drosophila) | - | HPRD | 9472028 |
| LBR | DHCR14B | FLJ43126 | LMN2R | MGC9041 | PHA | lamin B receptor | - | HPRD | 10049757 |10390541 |
| MBP | MGC99675 | myelin basic protein | - | HPRD | 10390541 |
| PRM1 | P1 | protamine 1 | - | HPRD,BioGRID | 10390541 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| SAFB | DKFZp779C1727 | HAP | HET | SAFB1 | scaffold attachment factor B | - | HPRD | 11509566 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | - | HPRD | 8208298 |8798720 |9472028 |10390541 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | Affinity Capture-Western Biochemical Activity Far Western Reconstituted Complex | BioGRID | 9472028 |10196197 |12565863 |17517895 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| SFRS2IP | CASP11 | SIP1 | SRRP129 | splicing factor, arginine/serine-rich 2, interacting protein | - | HPRD | 8208298 |
| SFRS3 | SRp20 | splicing factor, arginine/serine-rich 3 | - | HPRD | 8208298 |10390541 |
| SFRS4 | SRP75 | splicing factor, arginine/serine-rich 4 | - | HPRD | 8208298 |10390541 |
| SFRS5 | HRS | SRP40 | splicing factor, arginine/serine-rich 5 | - | HPRD | 8208298 |10390541 |
| SFRS6 | B52 | FLJ08061 | MGC5045 | SRP55 | splicing factor, arginine/serine-rich 6 | - | HPRD | 8208298 |10340541 |
| SNRNP70 | RNPU1Z | RPU1 | SNRP70 | U170K | U1AP | U1RNP | small nuclear ribonucleoprotein 70kDa (U1) | Affinity Capture-Western Biochemical Activity | BioGRID | 9472028 |12417631 |
| U2AF1 | DKFZp313J1712 | FP793 | RN | RNU2AF1 | U2AF35 | U2AFBP | U2 small nuclear RNA auxiliary factor 1 | - | HPRD | 9472028 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | - | HPRD | 9472028 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 15324660 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE DN | 47 | 34 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH PROLIFERATION | 147 | 80 | All SZGR 2.0 genes in this pathway |
| SCHUHMACHER MYC TARGETS UP | 80 | 57 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| NUMATA CSF3 SIGNALING VIA STAT3 | 22 | 17 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCER KINOME RED | 16 | 11 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN | 50 | 35 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER WITH EPCAM UP | 53 | 25 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 1828 | 1835 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-139 | 1695 | 1701 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-15/16/195/424/497 | 1181 | 1188 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 1104 | 1110 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-182 | 221 | 227 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-186 | 2199 | 2205 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-26 | 1240 | 1246 | m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-485-5p | 251 | 257 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-503 | 1182 | 1188 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-9 | 223 | 229 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 221 | 227 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.