Gene Page: SYK
Summary ?
| GeneID | 6850 |
| Symbol | SYK |
| Synonyms | p72-Syk |
| Description | spleen tyrosine kinase |
| Reference | MIM:600085|HGNC:HGNC:11491|Ensembl:ENSG00000165025|HPRD:02514|Vega:OTTHUMG00000020200 |
| Gene type | protein-coding |
| Map location | 9q22 |
| Pascal p-value | 0.501 |
| Sherlock p-value | 0.145 |
| Fetal beta | -0.402 |
| DMG | 1 (# studies) |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4032 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07160163 | 9 | 93563778 | SYK | 3.43E-8 | -0.014 | 1.02E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
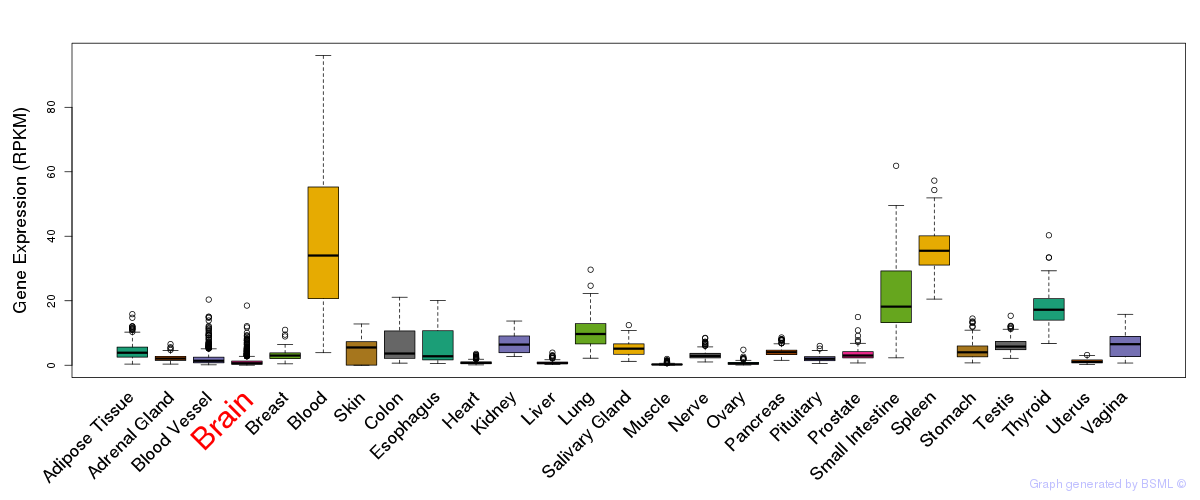
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IRF9 | 0.93 | 0.87 |
| PARP9 | 0.89 | 0.81 |
| IFITM3 | 0.88 | 0.79 |
| OAS1 | 0.88 | 0.75 |
| IFI35 | 0.87 | 0.81 |
| IFIT3 | 0.87 | 0.83 |
| PSMB8 | 0.87 | 0.83 |
| BATF2 | 0.87 | 0.78 |
| PLSCR1 | 0.87 | 0.74 |
| MT1E | 0.86 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RTF1 | -0.61 | -0.71 |
| PHF14 | -0.57 | -0.74 |
| NOL4 | -0.56 | -0.69 |
| DPF1 | -0.56 | -0.70 |
| PDCD11 | -0.56 | -0.66 |
| SH3BP5 | -0.55 | -0.66 |
| REV1 | -0.55 | -0.66 |
| RNASEN | -0.55 | -0.68 |
| SAFB | -0.55 | -0.66 |
| HEATR5B | -0.55 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004716 | receptor signaling protein tyrosine kinase activity | IEA | - | |
| GO:0005178 | integrin binding | IPI | 12885943 | |
| GO:0005515 | protein binding | IPI | 8627166 |9280292 |12885943 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001820 | serotonin secretion | IEA | neuron, serotonin (GO term level: 7) | - |
| GO:0006461 | protein complex assembly | TAS | 8163536 | |
| GO:0007257 | activation of JNK activity | IEA | - | |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0007229 | integrin-mediated signaling pathway | NAS | 12885943 | |
| GO:0009887 | organ morphogenesis | TAS | 7477352 | |
| GO:0007167 | enzyme linked receptor protein signaling pathway | IEA | - | |
| GO:0008283 | cell proliferation | TAS | 10963601 | |
| GO:0007159 | leukocyte adhesion | IDA | 12885943 | |
| GO:0019370 | leukotriene biosynthetic process | IEA | - | |
| GO:0030593 | neutrophil chemotaxis | IDA | 12885943 | |
| GO:0043366 | beta selection | IEA | - | |
| GO:0043306 | positive regulation of mast cell degranulation | IEA | - | |
| GO:0045579 | positive regulation of B cell differentiation | IEA | - | |
| GO:0045588 | positive regulation of gamma-delta T cell differentiation | IEA | - | |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IEA | - | |
| GO:0050850 | positive regulation of calcium-mediated signaling | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| GO:0046638 | positive regulation of alpha-beta T cell differentiation | IEA | - | |
| GO:0046641 | positive regulation of alpha-beta T cell proliferation | IEA | - | |
| GO:0046777 | protein amino acid autophosphorylation | IEA | - | |
| GO:0045401 | positive regulation of interleukin-3 biosynthetic process | IEA | - | |
| GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process | IEA | - | |
| GO:0050853 | B cell receptor signaling pathway | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019815 | B cell receptor complex | IEA | Synap (GO term level: 8) | - |
| GO:0042101 | T cell receptor complex | IDA | Synap (GO term level: 8) | 8176201 |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 9280292 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BLNK | BASH | BLNK-S | LY57 | MGC111051 | SLP-65 | SLP65 | B-cell linker | - | HPRD | 10981967 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Affinity Capture-Western Reconstituted Complex | BioGRID | 9857068 |11331248 |12435267 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD | 9857068 |10540342 |11133830 |
| CD19 | B4 | MGC12802 | CD19 molecule | - | HPRD,BioGRID | 9120258 |
| CD22 | FLJ22814 | MGC130020 | SIGLEC-2 | SIGLEC2 | CD22 molecule | - | HPRD,BioGRID | 8627166 |
| CD3E | FLJ18683 | T3E | TCRE | CD3e molecule, epsilon (CD3-TCR complex) | Reconstituted Complex | BioGRID | 7761456 |
| CD72 | CD72b | LYB2 | CD72 molecule | Biochemical Activity | BioGRID | 9590210 |
| CD79A | IGA | MB-1 | CD79a molecule, immunoglobulin-associated alpha | - | HPRD,BioGRID | 1569106|7500027 |
| CD79B | B29 | IGB | CD79b molecule, immunoglobulin-associated beta | Reconstituted Complex | BioGRID | 7500027 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | CrkL interacts with Syk. | BIND | 11313252 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD,BioGRID | 11313252 |
| CSF3R | CD114 | GCSFR | colony stimulating factor 3 receptor (granulocyte) | - | HPRD,BioGRID | 8197119 |
| CTTN | EMS1 | FLJ34459 | cortactin | - | HPRD,BioGRID | 8636079 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD,BioGRID | 9852052 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Syk interacts with ezrin. This interaction was modeled on a demonstrated interaction between Syk and ezrin from unspecified species. | BIND | 12387735 |
| FCER1G | FCRG | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | - | HPRD,BioGRID | 8071371 |
| FCGR1A | CD64 | CD64A | FCRI | FLJ18345 | IGFR1 | Fc fragment of IgG, high affinity Ia, receptor (CD64) | - | HPRD,BioGRID | 11141335 |
| FCGR2A | CD32 | CD32A | CDw32 | FCG2 | FCGR2 | FCGR2A1 | FcGR | IGFR2 | MGC23887 | MGC30032 | Fc fragment of IgG, low affinity IIa, receptor (CD32) | Reconstituted Complex | BioGRID | 9268059 |
| FCRL3 | FCRH3 | IFGP3 | IRTA3 | SPAP2 | Fc receptor-like 3 | Reconstituted Complex | BioGRID | 12051764 |
| FGR | FLJ43153 | MGC75096 | SRC2 | c-fgr | c-src2 | p55c-fgr | p58c-fgr | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | - | HPRD,BioGRID | 11672534 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 9535867 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 11964172 |
| ITGB2 | CD18 | LAD | LCAMB | LFA-1 | MAC-1 | MF17 | MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | Affinity Capture-Western | BioGRID | 10961871 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD | 10825200 |
| LAT | LAT1 | pp36 | linker for activation of T cells | Biochemical Activity | BioGRID | 11368773 |
| LAT | LAT1 | pp36 | linker for activation of T cells | - | HPRD | 9891970 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 7539035 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | - | HPRD | 15388330 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 7831290 |
| MS4A2 | APY | ATOPY | FCER1B | FCERI | IGEL | IGER | IGHER | MS4A1 | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | - | HPRD,BioGRID | 8071371 |
| MSN | - | moesin | Syk interacts with moesin. This interaction was modeled on a demonstrated interaction between Syk and moesin from unspecified species. | BIND | 12387735 |
| NFAM1 | CNAIP | FLJ40652 | bK126B4.4 | NFAT activating protein with ITAM motif 1 | - | HPRD | 15143214 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD,BioGRID | 10790433 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Affinity Capture-Western | BioGRID | 7831290 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD | 8885868 |
| PLCG2 | - | phospholipase C, gamma 2 (phosphatidylinositol-specific) | Affinity Capture-Western | BioGRID | 10981967 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | Affinity Capture-Western | BioGRID | 8885868 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 9169439 |9342235 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10747947 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | Affinity Capture-Western Biochemical Activity | BioGRID | 10072516 |10747947 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD | 10072516 |11356834 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 9590268 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Biochemical Activity | BioGRID | 16640565 |
| RPS6KB2 | KLS | P70-beta | P70-beta-1 | P70-beta-2 | S6K-beta2 | S6K2 | SRK | STK14B | p70(S6K)-beta | p70S6Kb | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | Biochemical Activity | BioGRID | 16640565 |
| SH2B2 | APS | SH2B adaptor protein 2 | - | HPRD | 10872802 |
| SH3BP2 | 3BP2 | CRBM | CRPM | FLJ42079 | RES4-23 | SH3-domain binding protein 2 | - | HPRD,BioGRID | 9846481 |11390470 |
| SIRPB1 | CD172b | DKFZp686A05192 | FLJ26614 | SIRP-BETA-1 | signal-regulatory protein beta 1 | Affinity Capture-Western | BioGRID | 10940905 |
| SIT1 | MGC125908 | MGC125909 | MGC125910 | RP11-331F9.5 | SIT | signaling threshold regulating transmembrane adaptor 1 | - | HPRD,BioGRID | 10209036 |
| SLA | SLA1 | SLAP | Src-like-adaptor | - | HPRD,BioGRID | 10449770 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 7513017 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | Syk interacts with and phosphorylates STAT1-alpha. | BIND | 15467722 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | Syk interacts with STAT1-beta. | BIND | 15467722 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD | 10825200 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD | 10825200 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | SYK autophosphorylates. | BIND | 12477728 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 9698567 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 11418661 |
| TUBA1A | B-ALPHA-1 | FLJ25113 | LIS3 | TUBA3 | tubulin, alpha 1a | - | HPRD,BioGRID | 10862713 |
| UBB | FLJ25987 | MGC8385 | ubiquitin B | Syk interacts with ubiquitin. | BIND | 11046148 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 8986718 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPPA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2 PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKCELLS PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| SA B CELL RECEPTOR COMPLEXES | 24 | 20 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID NFKAPPAB ATYPICAL PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF SIGNALING BY CBL | 18 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 2 SIGNALING | 41 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS DN | 81 | 42 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 8 | 18 | 7 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 DN | 64 | 47 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA DN | 41 | 27 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE SULINDAC 4 | 21 | 12 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CLASPER LYMPHATIC VESSELS DURING METASTASIS DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY 4NQO IN OLD | 13 | 10 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY 4NQO IN WS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN WS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE DN | 21 | 12 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCER KINOME GREEN | 16 | 14 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER PROGRESSION UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE UP | 107 | 59 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ WALDENSTROEMS MACROGLOBULINEMIA 2 | 13 | 9 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |