Gene Page: C1QBP
Summary ?
| GeneID | 708 |
| Symbol | C1QBP |
| Synonyms | GC1QBP|HABP1|SF2p32|gC1Q-R|gC1qR|p32 |
| Description | complement component 1, q subcomponent binding protein |
| Reference | MIM:601269|HGNC:HGNC:1243|Ensembl:ENSG00000108561|HPRD:03168|Vega:OTTHUMG00000177875 |
| Gene type | protein-coding |
| Map location | 17p13.3 |
| Pascal p-value | 0.023 |
| Sherlock p-value | 0.662 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_clathrin G2Cdb.human_mitochondria G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06345277 | 17 | 5342595 | C1QBP | 1.966E-4 | -0.229 | 0.035 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
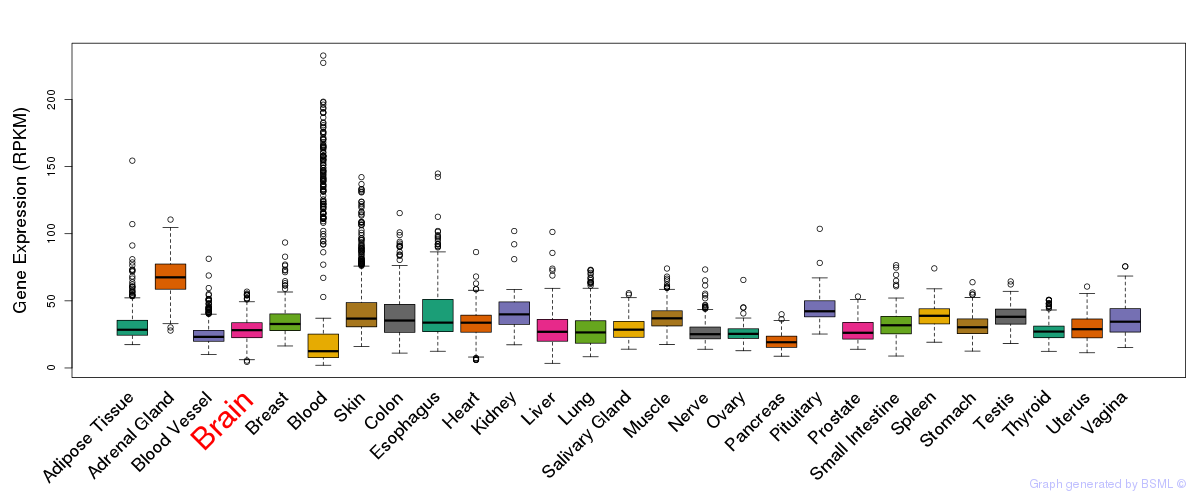
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1QB | 0.89 | 0.94 |
| C1QC | 0.89 | 0.87 |
| TYROBP | 0.88 | 0.91 |
| RNASE6 | 0.87 | 0.84 |
| TBXAS1 | 0.81 | 0.82 |
| FCER1G | 0.81 | 0.86 |
| ADORA3 | 0.80 | 0.78 |
| GPR34 | 0.79 | 0.77 |
| SLC2A5 | 0.79 | 0.73 |
| MS4A6A | 0.78 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UPF3A | -0.48 | -0.66 |
| THOC2 | -0.48 | -0.60 |
| SAFB2 | -0.48 | -0.55 |
| MAP4K4 | -0.47 | -0.59 |
| CDC2L2 | -0.47 | -0.53 |
| UPF3B | -0.47 | -0.63 |
| ZNF418 | -0.47 | -0.54 |
| RTF1 | -0.46 | -0.53 |
| SFRS8 | -0.46 | -0.55 |
| ZC3H13 | -0.46 | -0.51 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT2 | D6S51 | D6S51E | DKFZp686D09175 | G2 | HLA-B associated transcript 2 | - | HPRD,BioGRID | 14667819 |
| CBFB | PEBP2B | core-binding factor, beta subunit | p32 interacts with CBF-B. | BIND | 15243141 |
| EXOSC6 | EAP4 | MTR3 | Mtr3p | hMtr3p | p11 | exosome component 6 | - | HPRD | 15231747 |
| FBL | FIB | FLRN | RNU3IP1 | fibrillarin | Affinity Capture-MS | BioGRID | 14583623 |
| GAB1 | - | GRB2-associated binding protein 1 | - | HPRD,BioGRID | 10747014 |
| GABRB1 | - | gamma-aminobutyric acid (GABA) A receptor, beta 1 | - | HPRD | 11350968 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | - | HPRD | 14743216 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD,BioGRID | 11866440 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD,BioGRID | 11866440 |
| MMP14 | MMP-X1 | MT1-MMP | MTMMP1 | matrix metallopeptidase 14 (membrane-inserted) | - | HPRD,BioGRID | 12220632 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD,BioGRID | 10831594 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | - | HPRD,BioGRID | 10831594 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD,BioGRID | 10831594 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | - | HPRD,BioGRID | 10831594 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | - | HPRD,BioGRID | 10022843 |
| SFRS9 | SRp30c | splicing factor, arginine/serine-rich 9 | Far Western | BioGRID | 10022843 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN DN | 27 | 20 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| COLLER MYC TARGETS UP | 25 | 19 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI DN | 73 | 45 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 UP | 65 | 44 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS UP | 47 | 31 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA DN | 100 | 63 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| IRITANI MAD1 TARGETS DN | 47 | 30 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL CUTANEOUS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |