Gene Page: VARS
Summary ?
| GeneID | 7407 |
| Symbol | VARS |
| Synonyms | G7A|VARS1|VARS2 |
| Description | valyl-tRNA synthetase |
| Reference | MIM:192150|HGNC:HGNC:12651|Ensembl:ENSG00000204394|HPRD:10362|Vega:OTTHUMG00000031286 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1E-12 |
| Fetal beta | 0.611 |
| eGene | Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0113 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
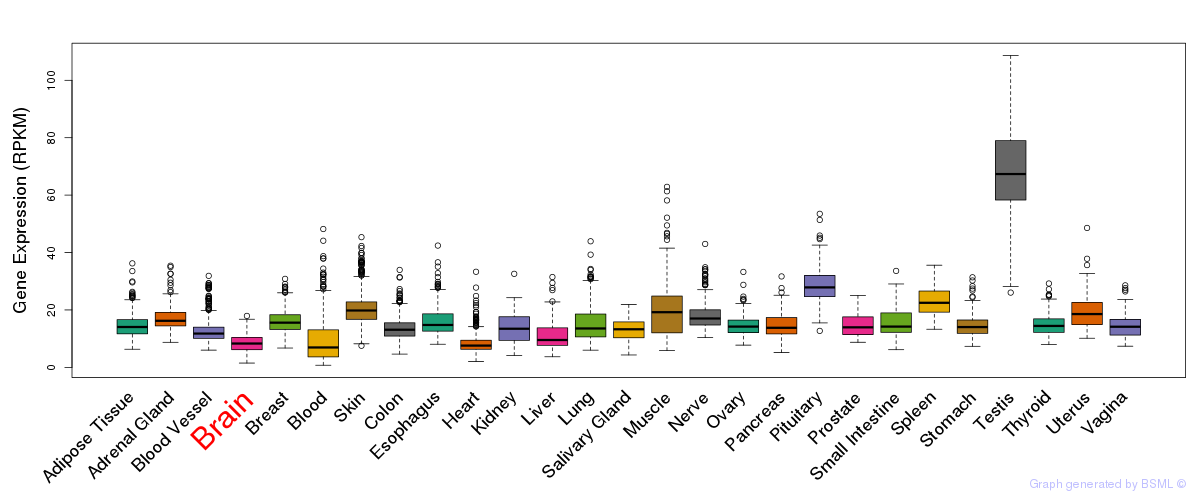
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLCO2B1 | 0.83 | 0.84 |
| MMRN2 | 0.83 | 0.87 |
| A2M | 0.78 | 0.79 |
| ITIH5 | 0.77 | 0.83 |
| TNFRSF1B | 0.77 | 0.78 |
| CDH5 | 0.74 | 0.80 |
| BGN | 0.74 | 0.76 |
| ENG | 0.74 | 0.80 |
| CLEC14A | 0.74 | 0.75 |
| LRRC32 | 0.73 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF32 | -0.52 | -0.58 |
| C8orf59 | -0.51 | -0.59 |
| SNHG12 | -0.51 | -0.56 |
| OXSM | -0.50 | -0.57 |
| NDUFAF2 | -0.50 | -0.56 |
| C12orf45 | -0.49 | -0.58 |
| ST20 | -0.49 | -0.55 |
| MTIF3 | -0.47 | -0.52 |
| ATP5E | -0.47 | -0.60 |
| MRPL21 | -0.46 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004832 | valine-tRNA ligase activity | IDA | 8428657 | |
| GO:0004832 | valine-tRNA ligase activity | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006414 | translational elongation | NAS | - | |
| GO:0006438 | valyl-tRNA aminoacylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | NAS | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | - | HPRD,BioGRID | 2556394 |3169261 |
| EEF1A2 | EEF1AL | EF-1-alpha-2 | EF1A | FLJ41696 | HS1 | STN | STNL | eukaryotic translation elongation factor 1 alpha 2 | Affinity Capture-MS | BioGRID | 17353931 |
| EEF1B2 | EEF1B | EEF1B1 | EF1B | eukaryotic translation elongation factor 1 beta 2 | - | HPRD,BioGRID | 2556394 |3169261 |
| EEF1D | EF-1D | EF1D | FLJ20897 | FP1047 | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | - | HPRD | 2556394 |3169261 |
| EEF1D | EF-1D | EF1D | FLJ20897 | FP1047 | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | - | HPRD,BioGRID | 2556394 |3169261|8294461 |
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Co-purification | BioGRID | 3169261 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | Affinity Capture-MS | BioGRID | 17353931 |
| GFM1 | COXPD1 | EFG | EFG1 | EFGM | EGF1 | FLJ12662 | FLJ13632 | FLJ20773 | GFM | hEFG1 | G elongation factor, mitochondrial 1 | - | HPRD | 2556394 |3169261 |
| HLA-B | AS | HLA-B-7301 | HLA-B73 | HLAB | HLAC | SPDA1 | major histocompatibility complex, class I, B | Affinity Capture-MS | BioGRID | 17353931 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | - | HPRD | 14743216 |
| TFE3 | RCCP2 | TFEA | bHLHe33 | transcription factor binding to IGHM enhancer 3 | Affinity Capture-MS | BioGRID | 17353931 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VALINE LEUCINE AND ISOLEUCINE BIOSYNTHESIS | 11 | 8 | All SZGR 2.0 genes in this pathway |
| KEGG AMINOACYL TRNA BIOSYNTHESIS | 41 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOSOLIC TRNA AMINOACYLATION | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRNA AMINOACYLATION | 42 | 34 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| CEBALLOS TARGETS OF TP53 AND MYC DN | 38 | 31 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| SCHUHMACHER MYC TARGETS UP | 80 | 57 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA UP | 51 | 29 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 30MIN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 52 | 58 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.