Gene Page: WRN
Summary ?
| GeneID | 7486 |
| Symbol | WRN |
| Synonyms | RECQ3|RECQL2|RECQL3 |
| Description | Werner syndrome RecQ like helicase |
| Reference | MIM:604611|HGNC:HGNC:12791|Ensembl:ENSG00000165392|HPRD:05212|Vega:OTTHUMG00000163894 |
| Gene type | protein-coding |
| Map location | 8p12 |
| Pascal p-value | 0.022 |
| Fetal beta | 0.947 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WRN | chr8 | 31004622 | C | T | NM_000553 | p.1146S>L | missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10709021 | 8 | 30890108 | PURG;WRN | 1.583E-4 | 0.432 | 0.032 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
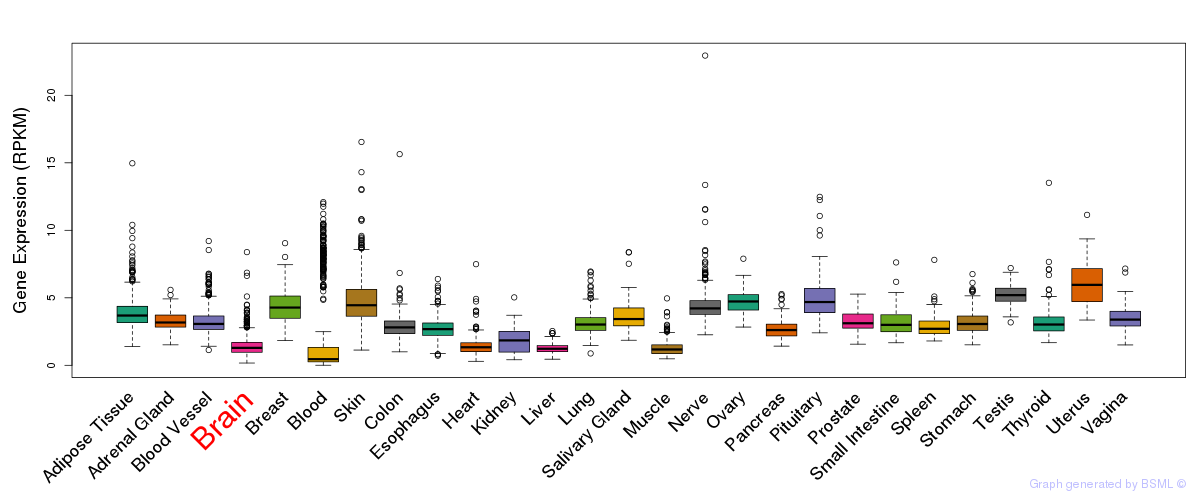
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB6A | 0.93 | 0.91 |
| ITFG1 | 0.92 | 0.92 |
| ARMCX3 | 0.92 | 0.90 |
| CLTC | 0.92 | 0.93 |
| C5orf22 | 0.91 | 0.92 |
| USP14 | 0.91 | 0.89 |
| VPS41 | 0.91 | 0.88 |
| PNMA2 | 0.91 | 0.89 |
| SRPK2 | 0.91 | 0.91 |
| NCKAP1 | 0.90 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.65 | -0.52 |
| AF347015.2 | -0.65 | -0.53 |
| MT-CO2 | -0.64 | -0.54 |
| EIF4EBP3 | -0.63 | -0.65 |
| AF347015.8 | -0.63 | -0.53 |
| HIGD1B | -0.63 | -0.54 |
| FXYD1 | -0.62 | -0.55 |
| AF347015.31 | -0.61 | -0.53 |
| AF347015.26 | -0.61 | -0.49 |
| MT-CYB | -0.60 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000403 | Y-form DNA binding | IDA | 17715146 | |
| GO:0000405 | bubble DNA binding | IDA | 11433031 | |
| GO:0003677 | DNA binding | IDA | 9288107 | |
| GO:0004003 | ATP-dependent DNA helicase activity | IDA | 9288107 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008408 | 3'-5' exonuclease activity | IDA | 10783163 |12181313 | |
| GO:0009378 | four-way junction helicase activity | IDA | 11433031 | |
| GO:0016887 | ATPase activity | IDA | 10373438 |10871376 | |
| GO:0042803 | protein homodimerization activity | IDA | 10783163 | |
| GO:0032403 | protein complex binding | IDA | 10783163 | |
| GO:0043138 | 3'-5' DNA helicase activity | IDA | 17715146 | |
| GO:0051880 | G-quadruplex DNA binding | IDA | 11433031 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000723 | telomere maintenance | IMP | 18212065 | |
| GO:0000731 | DNA synthesis during DNA repair | IDA | 17563354 | |
| GO:0001302 | replicative cell aging | IEA | - | |
| GO:0006310 | DNA recombination | IEA | - | |
| GO:0006284 | base-excision repair | IDA | 17611195 | |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | IEA | - | |
| GO:0010225 | response to UV-C | IDA | 17563354 | |
| GO:0010259 | multicellular organismal aging | IMP | 16673358 | |
| GO:0006979 | response to oxidative stress | IDA | 17611195 | |
| GO:0042981 | regulation of apoptosis | IGI | 9681877 | |
| GO:0051345 | positive regulation of hydrolase activity | IDA | 17611195 | |
| GO:0031297 | replication fork processing | IDA | 17115688 | |
| GO:0031297 | replication fork processing | IMP | 12882351 | |
| GO:0040009 | regulation of growth rate | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IDA | 17498979 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | IDA | 11420665 |12944467 | |
| GO:0005730 | nucleolus | IDA | 9618508 |12181313 | |
| GO:0032389 | MutLalpha complex | IDA | 17715146 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Protein-peptide | BioGRID | 10608806 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Protein-peptide | BioGRID | 10608806 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | BLM interacts with WRN. | BIND | 11919194 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | - | HPRD,BioGRID | 11919194 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | p14 interacts with WRN. | BIND | 15355988 |
| FEN1 | FEN-1 | MF1 | RAD2 | flap structure-specific endonuclease 1 | - | HPRD | 12356323 |
| FEN1 | FEN-1 | MF1 | RAD2 | flap structure-specific endonuclease 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11598021 |14688284 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | Protein-peptide | BioGRID | 14578343 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | WRN interacts with PARP1. | BIND | 14734561 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 10871373 |12633936 |
| POLR1C | RPA39 | RPA40 | RPA5 | RPAC1 | polymerase (RNA) I polypeptide C, 30kDa | - | HPRD,BioGRID | 11971179 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD,BioGRID | 11889123 |
| RAD52 | - | RAD52 homolog (S. cerevisiae) | WRN interacts with RAD52. | BIND | 12750383 |
| RDM1 | MGC33977 | RAD52B | RAD52 motif 1 | - | HPRD | 12750383 |
| TERF2 | TRBF2 | TRF2 | telomeric repeat binding factor 2 | - | HPRD,BioGRID | 12181313 |
| TERF2 | TRBF2 | TRF2 | telomeric repeat binding factor 2 | Wrn interacts with TRF2. | BIND | 12181313 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | WRN interacts with p53. | BIND | 12080066 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11427532 |12080066 |
| WRNIP1 | FLJ22526 | RP11-420G6.2 | WHIP | bA420G6.2 | Werner helicase interacting protein 1 | - | HPRD,BioGRID | 11301316 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | - | HPRD,BioGRID | 12177300 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | WRN interacts with G22P1 (Ku70) as part of a complex. | BIND | 14734561 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | - | HPRD,BioGRID | 12177300 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| MARKS ACETYLATED NON HISTONE PROTEINS | 15 | 9 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 9 | 8 | 6 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP | 81 | 57 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 DN | 64 | 47 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |