Gene Page: NUP37
Summary ?
| GeneID | 79023 |
| Symbol | NUP37 |
| Synonyms | p37 |
| Description | nucleoporin 37kDa |
| Reference | MIM:609264|HGNC:HGNC:29929|Ensembl:ENSG00000075188|HPRD:14853| |
| Gene type | protein-coding |
| Map location | 12q23.2 |
| Pascal p-value | 0.877 |
| Fetal beta | 0.5 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NUP37 | chr12 | 102512191 | T | C | NM_024057 | p.36I>V | missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
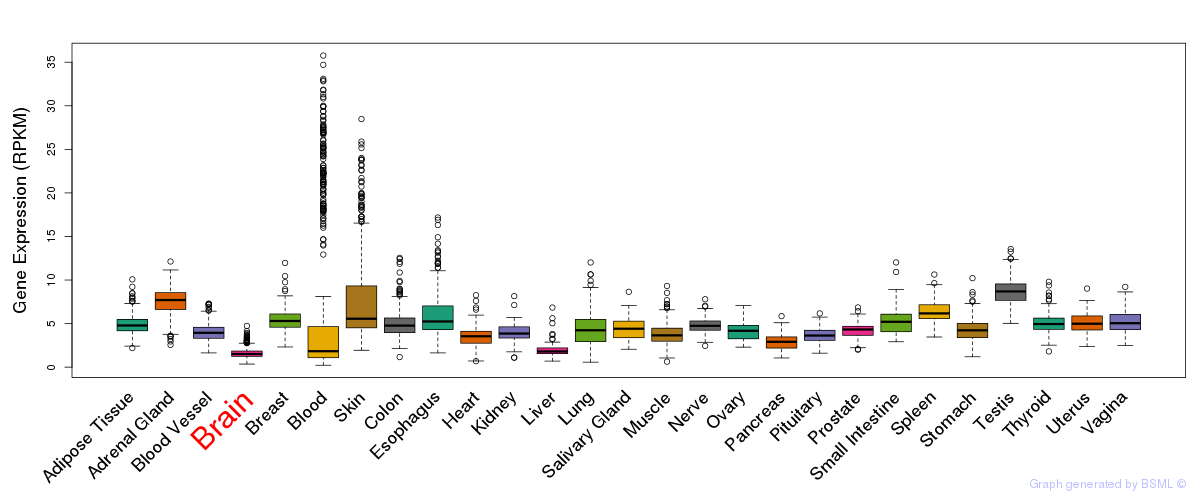
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME METABOLISM OF NON CODING RNA | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE TRANSPORT | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |