Gene Page: DHRS12
Summary ?
| GeneID | 79758 |
| Symbol | DHRS12 |
| Synonyms | SDR40C1 |
| Description | dehydrogenase/reductase (SDR family) member 12 |
| Reference | MIM:616163|HGNC:HGNC:25832|Ensembl:ENSG00000102796|HPRD:07833|Vega:OTTHUMG00000016952 |
| Gene type | protein-coding |
| Map location | 13q14.3 |
| Pascal p-value | 0.372 |
| Sherlock p-value | 0.131 |
| Fetal beta | -0.372 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26851608 | 13 | 52378480 | DHRS12 | 2.204E-4 | -0.348 | 0.036 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3990008 | 13 | 52331598 | DHRS12 | ENSG00000102796.6 | 1.93419E-6 | 0.01 | 46695 | gtex_brain_ba24 |
| rs57280031 | 13 | 52345781 | DHRS12 | ENSG00000102796.6 | 1.72492E-7 | 0.01 | 32512 | gtex_brain_ba24 |
| rs7985214 | 13 | 52347439 | DHRS12 | ENSG00000102796.6 | 1.49848E-7 | 0.01 | 30854 | gtex_brain_ba24 |
| rs9596573 | 13 | 52347553 | DHRS12 | ENSG00000102796.6 | 1.51992E-7 | 0.01 | 30740 | gtex_brain_ba24 |
| rs57280031 | 13 | 52345781 | DHRS12 | ENSG00000102796.6 | 3.96373E-6 | 0.04 | 32512 | gtex_brain_putamen_basal |
| rs7985214 | 13 | 52347439 | DHRS12 | ENSG00000102796.6 | 3.58743E-6 | 0.04 | 30854 | gtex_brain_putamen_basal |
| rs9596573 | 13 | 52347553 | DHRS12 | ENSG00000102796.6 | 3.65318E-6 | 0.04 | 30740 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
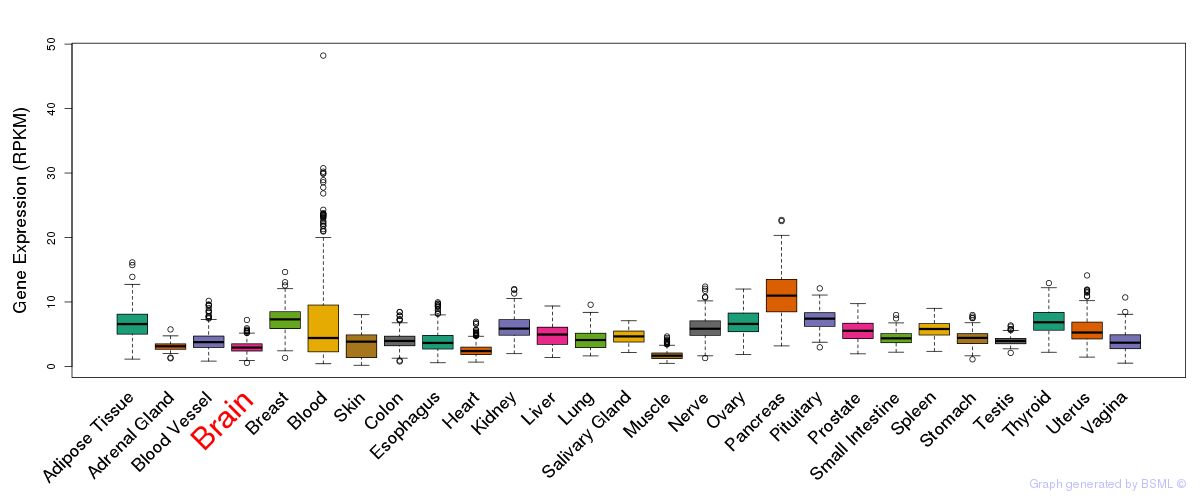
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 UP | 125 | 61 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 2 UP | 54 | 31 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |