Gene Page: GLRA3
Summary ?
| GeneID | 8001 |
| Symbol | GLRA3 |
| Synonyms | - |
| Description | glycine receptor alpha 3 |
| Reference | MIM:600421|HGNC:HGNC:4328|Ensembl:ENSG00000145451|HPRD:02688|Vega:OTTHUMG00000149816 |
| Gene type | protein-coding |
| Map location | 4q34.1 |
| Pascal p-value | 0.732 |
| DMG | 1 (# studies) |
| eGene | Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09841488 | 4 | 175750527 | GLRA3 | 2.667E-4 | -0.236 | 0.038 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs695809 | chr22 | 26278127 | GLRA3 | 8001 | 0.05 | trans | ||
| rs330478 | 4 | 176050287 | GLRA3 | ENSG00000145451.8 | 2.10962E-6 | 0.02 | -299822 | gtex_brain_putamen_basal |
| rs5864311 | 4 | 176108113 | GLRA3 | ENSG00000145451.8 | 2.54893E-6 | 0.02 | -357648 | gtex_brain_putamen_basal |
| rs1491416 | 4 | 176109168 | GLRA3 | ENSG00000145451.8 | 8.63185E-7 | 0.02 | -358703 | gtex_brain_putamen_basal |
| rs1121867 | 4 | 176111188 | GLRA3 | ENSG00000145451.8 | 2.89645E-6 | 0.02 | -360723 | gtex_brain_putamen_basal |
| rs12641212 | 4 | 176114955 | GLRA3 | ENSG00000145451.8 | 8.8121E-7 | 0.02 | -364490 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
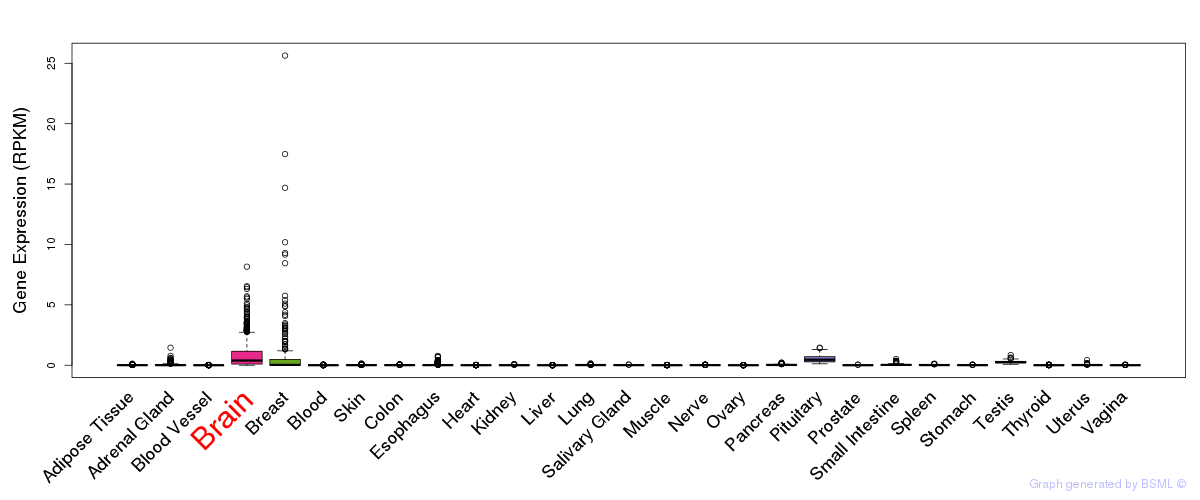
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WARS | 0.91 | 0.91 |
| ITFG1 | 0.90 | 0.88 |
| SUCLA2 | 0.90 | 0.88 |
| PRKAR1A | 0.90 | 0.91 |
| ATP2A2 | 0.89 | 0.90 |
| AP3M2 | 0.89 | 0.89 |
| PLEKHB2 | 0.89 | 0.92 |
| YWHAB | 0.89 | 0.87 |
| ARL8B | 0.89 | 0.91 |
| NDUFS1 | 0.89 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.59 | -0.37 |
| AF347015.18 | -0.56 | -0.40 |
| AP002478.3 | -0.54 | -0.46 |
| AF347015.2 | -0.53 | -0.34 |
| MT-CO2 | -0.53 | -0.38 |
| HIGD1B | -0.52 | -0.38 |
| AC098691.2 | -0.52 | -0.41 |
| AF347015.8 | -0.52 | -0.37 |
| CSAG1 | -0.51 | -0.39 |
| AL139819.3 | -0.51 | -0.43 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030594 | neurotransmitter receptor activity | IEA | Neurotransmitter (GO term level: 5) | - |
| GO:0004890 | GABA-A receptor activity | IEA | GABA (GO term level: 7) | - |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | NAS | - | |
| GO:0016934 | extracellular-glycine-gated chloride channel activity | IEA | - | |
| GO:0031404 | chloride ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | NAS | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0006821 | chloride transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0016021 | integral to membrane | NAS | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME LIGAND GATED ION CHANNEL TRANSPORT | 21 | 21 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| MCCABE HOXC6 TARGETS CANCER UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |