Gene Page: KHSRP
Summary ?
| GeneID | 8570 |
| Symbol | KHSRP |
| Synonyms | FBP2|FUBP2|KSRP |
| Description | KH-type splicing regulatory protein |
| Reference | MIM:603445|HGNC:HGNC:6316|Ensembl:ENSG00000088247|HPRD:10350|Vega:OTTHUMG00000180850 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.395 |
| Sherlock p-value | 0.145 |
| Fetal beta | 0.845 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08340521 | 19 | 6424783 | KHSRP | 9.18E-6 | -0.274 | 0.013 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
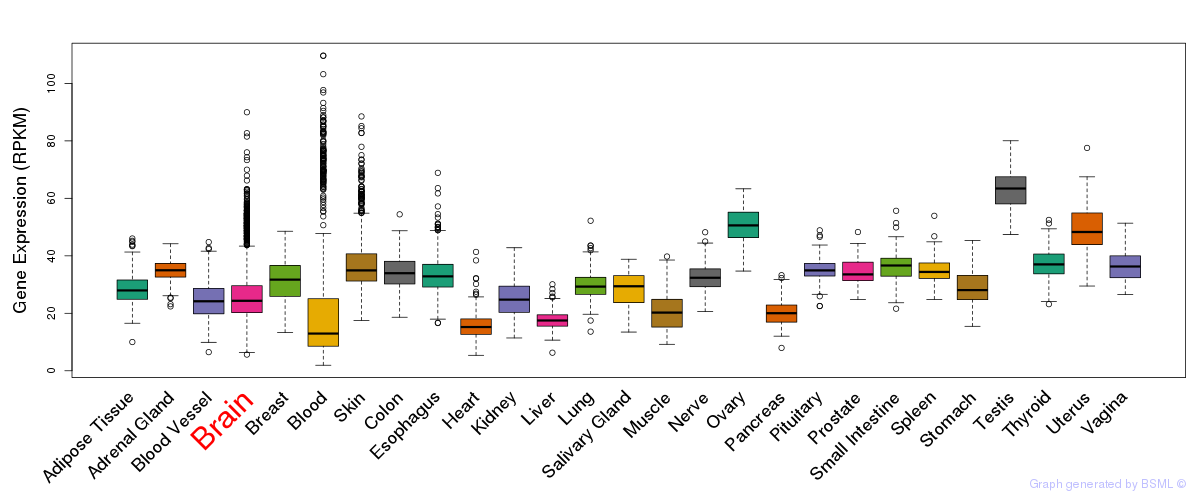
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COL14A1 | 0.82 | 0.86 |
| ACTA2 | 0.80 | 0.85 |
| FAM129A | 0.80 | 0.77 |
| JPH2 | 0.75 | 0.79 |
| MYH11 | 0.75 | 0.80 |
| MRGPRF | 0.71 | 0.78 |
| COL8A1 | 0.71 | 0.80 |
| C7orf58 | 0.70 | 0.60 |
| TBX18 | 0.68 | 0.74 |
| FHL5 | 0.66 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RP9 | -0.45 | -0.56 |
| ZNF433 | -0.44 | -0.53 |
| CYP2R1 | -0.43 | -0.56 |
| CCDC23 | -0.43 | -0.54 |
| RPAIN | -0.43 | -0.52 |
| TRNAU1AP | -0.42 | -0.52 |
| TUBB2B | -0.42 | -0.42 |
| RPS13P2 | -0.42 | -0.60 |
| DYNLT1 | -0.42 | -0.60 |
| RPL23A | -0.42 | -0.57 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| A1CF | ACF | ACF64 | ACF65 | APOBEC1CF | ASP | MGC163391 | RP11-564C4.2 | APOBEC1 complementation factor | - | HPRD,BioGRID | 10781591 |
| BACE1 | ASP2 | BACE | FLJ90568 | HSPC104 | KIAA1149 | beta-site APP-cleaving enzyme 1 | BACE1 interacts with KHSRP. This interaction was modelled on a demonstrated interaction between mouse BACE1 and human KHSRP. | BIND | 15606899 |
| EXOSC2 | RRP4 | Rrp4p | hRrp4p | p7 | exosome component 2 | KSRP interacts with hRrp4p. | BIND | 15175153 |
| EXOSC2 | RRP4 | Rrp4p | hRrp4p | p7 | exosome component 2 | - | HPRD,BioGRID | 15175153 |
| EXOSC3 | CGI-102 | MGC15120 | MGC723 | RP11-3J10.8 | RRP40 | Rrp40p | bA3J10.7 | hRrp40p | p10 | exosome component 3 | - | HPRD | 11719186 |15175153|15175153 |
| EXOSC3 | CGI-102 | MGC15120 | MGC723 | RP11-3J10.8 | RRP40 | Rrp40p | bA3J10.7 | hRrp40p | p10 | exosome component 3 | KSRP interacts with hRrp40p. | BIND | 15175153 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | KSRP interacts with AREfos. This interaction was modeled on a demonstrated interaction between KSRP from human and AREfos from an unspecified species. | BIND | 15175153 |
| FUBP1 | FBP | FUBP | far upstream element (FUSE) binding protein 1 | - | HPRD | 11003644 |
| HNRNPA1 | HNRPA1 | MGC102835 | heterogeneous nuclear ribonucleoprotein A1 | Reconstituted Complex | BioGRID | 11003644 |
| HNRNPF | HNRPF | MGC110997 | OK/SW-cl.23 | mcs94-1 | heterogeneous nuclear ribonucleoprotein F | - | HPRD | 9858532 |
| HNRNPH1 | DKFZp686A15170 | HNRPH | HNRPH1 | hnRNPH | heterogeneous nuclear ribonucleoprotein H1 (H) | - | HPRD,BioGRID | 9858532 |11003644 |
| IL2 | IL-2 | TCGF | lymphokine | interleukin 2 | KSRP interacts with IL-2 3'UTR. This interaction was modeled on a demonstrated interaction between KSRP from human and IL-2 3'UTR from an unspecified species. | BIND | 15175153 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | KSRP interacts with AREjun. This interaction was modeled on a demonstrated interaction between KSRP from human and AREjun from an unspecified species. | BIND | 15175153 |
| PARN | DAN | poly(A)-specific ribonuclease (deadenylation nuclease) | - | HPRD,BioGRID | 15175153 |
| PTBP1 | HNRNP-I | HNRNPI | HNRPI | MGC10830 | MGC8461 | PTB | PTB-1 | PTB-T | PTB2 | PTB3 | PTB4 | pPTB | polypyrimidine tract binding protein 1 | - | HPRD | 11003644 |
| PTBP2 | FLJ34897 | PTB | PTBLP | brPTB | nPTB | nPTB5 | nPTB6 | nPTB7 | nPTB8 | polypyrimidine tract binding protein 2 | Reconstituted Complex | BioGRID | 11003644 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Affinity Capture-MS | BioGRID | 17353931 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | KSRP interacts with AREtnf. This interaction was modeled on a demonstrated interaction between KSRP from human and AREtnf from an unspecified species. | BIND | 15175153 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PERK REGULATED GENE EXPRESSION | 29 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF GENES BY ATF4 | 26 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY KSRP | 17 | 9 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP DN | 11 | 8 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC AND SERUM RESPONSE SYNERGY | 32 | 22 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 24HR 5 UP | 34 | 23 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| PETRETTO BLOOD PRESSURE UP | 12 | 8 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BACOLOD RESISTANCE TO ALKYLATING AGENTS DN | 60 | 45 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING UP | 59 | 44 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM RESPONSE TRANSLATION | 37 | 20 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |