Gene Page: GGH
Summary ?
| GeneID | 8836 |
| Symbol | GGH |
| Synonyms | GH |
| Description | gamma-glutamyl hydrolase |
| Reference | MIM:601509|HGNC:HGNC:4248|Ensembl:ENSG00000137563|HPRD:03299|Vega:OTTHUMG00000164365 |
| Gene type | protein-coding |
| Map location | 8q12.3 |
| Pascal p-value | 0.723 |
| Sherlock p-value | 0.426 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02212000 | 8 | 63951669 | GGH | 7.46E-9 | -0.01 | 3.68E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10504360 | 8 | 63862337 | GGH | ENSG00000137563.7 | 4.54E-6 | 0.02 | 89393 | gtex_brain_ba24 |
| rs113252212 | 8 | 63864411 | GGH | ENSG00000137563.7 | 4.54E-6 | 0.02 | 87319 | gtex_brain_ba24 |
| rs72656550 | 8 | 63879439 | GGH | ENSG00000137563.7 | 4.54E-6 | 0.02 | 72291 | gtex_brain_ba24 |
| rs397838493 | 8 | 63880282 | GGH | ENSG00000137563.7 | 4.528E-6 | 0.02 | 71448 | gtex_brain_ba24 |
| rs17192653 | 8 | 63881321 | GGH | ENSG00000137563.7 | 4.505E-6 | 0.02 | 70409 | gtex_brain_ba24 |
| rs62508088 | 8 | 63883970 | GGH | ENSG00000137563.7 | 4.54E-6 | 0.02 | 67760 | gtex_brain_ba24 |
| rs62508108 | 8 | 63889037 | GGH | ENSG00000137563.7 | 1.831E-6 | 0.02 | 62693 | gtex_brain_ba24 |
| rs112685605 | 8 | 63890832 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 60898 | gtex_brain_ba24 |
| rs62508116 | 8 | 63904617 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 47113 | gtex_brain_ba24 |
| rs16930060 | 8 | 63906199 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 45531 | gtex_brain_ba24 |
| rs16930066 | 8 | 63907144 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 44586 | gtex_brain_ba24 |
| rs62508123 | 8 | 63908088 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 43642 | gtex_brain_ba24 |
| rs61518338 | 8 | 63909596 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 42134 | gtex_brain_ba24 |
| rs16930072 | 8 | 63911296 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 40434 | gtex_brain_ba24 |
| rs61629446 | 8 | 63917641 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 34089 | gtex_brain_ba24 |
| rs62510060 | 8 | 63918764 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 32966 | gtex_brain_ba24 |
| rs17279320 | 8 | 63921513 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 30217 | gtex_brain_ba24 |
| rs17279355 | 8 | 63922402 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 29328 | gtex_brain_ba24 |
| rs62510061 | 8 | 63924414 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 27316 | gtex_brain_ba24 |
| rs62510062 | 8 | 63924428 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 27302 | gtex_brain_ba24 |
| rs62510063 | 8 | 63924705 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 27025 | gtex_brain_ba24 |
| rs62510064 | 8 | 63926622 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 25108 | gtex_brain_ba24 |
| rs17279558 | 8 | 63927746 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 23984 | gtex_brain_ba24 |
| rs72658344 | 8 | 63928087 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 23643 | gtex_brain_ba24 |
| rs55800505 | 8 | 63930559 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 21171 | gtex_brain_ba24 |
| rs148625779 | 8 | 63930927 | GGH | ENSG00000137563.7 | 4.488E-6 | 0.02 | 20803 | gtex_brain_ba24 |
| rs11545078 | 8 | 63938764 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 12966 | gtex_brain_ba24 |
| rs3780129 | 8 | 63940962 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 10768 | gtex_brain_ba24 |
| rs3780127 | 8 | 63941139 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 10591 | gtex_brain_ba24 |
| rs57776226 | 8 | 63941840 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 9890 | gtex_brain_ba24 |
| rs17194931 | 8 | 63944344 | GGH | ENSG00000137563.7 | 1.846E-6 | 0.02 | 7386 | gtex_brain_ba24 |
| rs59169978 | 8 | 63945258 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 6472 | gtex_brain_ba24 |
| rs60889737 | 8 | 63945267 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 6463 | gtex_brain_ba24 |
| rs11990678 | 8 | 63945773 | GGH | ENSG00000137563.7 | 4.499E-6 | 0.02 | 5957 | gtex_brain_ba24 |
Section II. Transcriptome annotation
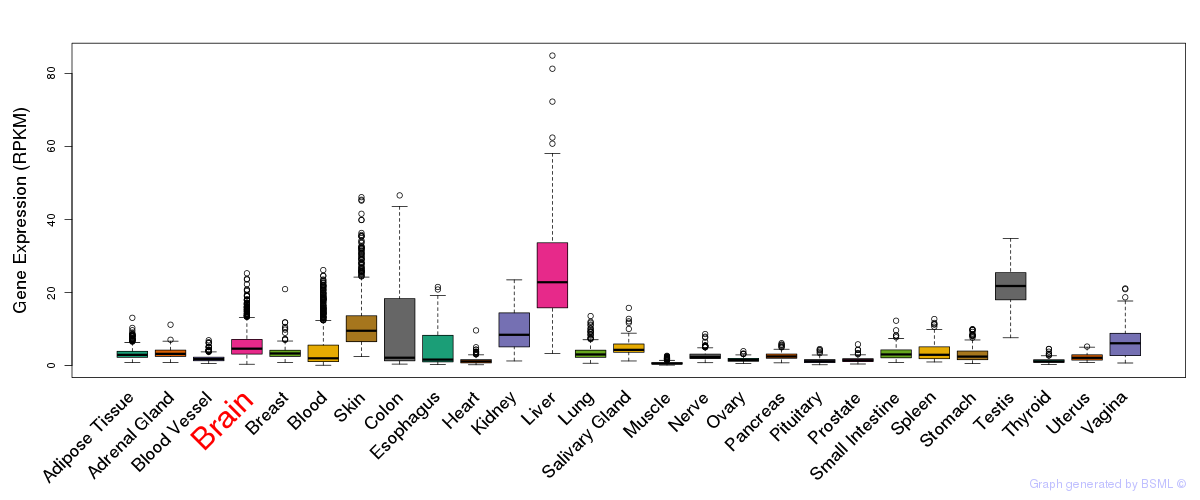
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMARCAD1 | 0.94 | 0.94 |
| TOP2B | 0.94 | 0.93 |
| NOL11 | 0.94 | 0.94 |
| DHX29 | 0.94 | 0.93 |
| GART | 0.94 | 0.91 |
| POLR2B | 0.94 | 0.92 |
| PTBP2 | 0.94 | 0.92 |
| ARFGAP3 | 0.94 | 0.92 |
| SENP1 | 0.93 | 0.92 |
| CEP57 | 0.93 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.75 | -0.80 |
| MT-CO2 | -0.74 | -0.88 |
| AF347015.31 | -0.73 | -0.86 |
| AIFM3 | -0.73 | -0.78 |
| AF347015.27 | -0.73 | -0.84 |
| AF347015.33 | -0.72 | -0.85 |
| FXYD1 | -0.72 | -0.86 |
| C5orf53 | -0.72 | -0.73 |
| MT-CYB | -0.71 | -0.85 |
| IFI27 | -0.71 | -0.85 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOLATE BIOSYNTHESIS | 11 | 10 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR DN | 88 | 53 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING DN | 87 | 49 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP | 69 | 40 | All SZGR 2.0 genes in this pathway |
| CAVARD LIVER CANCER MALIGNANT VS BENIGN | 32 | 19 | All SZGR 2.0 genes in this pathway |
| IWANAGA E2F1 TARGETS INDUCED BY SERUM | 31 | 19 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS DN | 121 | 65 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS UP | 48 | 36 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR | 74 | 47 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE UP | 107 | 59 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G6 UP | 65 | 43 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |