Gene Page: ROCK2
Summary ?
| GeneID | 9475 |
| Symbol | ROCK2 |
| Synonyms | ROCK-II |
| Description | Rho associated coiled-coil containing protein kinase 2 |
| Reference | MIM:604002|HGNC:HGNC:10252|Ensembl:ENSG00000134318|HPRD:04922|Vega:OTTHUMG00000149916 |
| Gene type | protein-coding |
| Map location | 2p24 |
| Pascal p-value | 0.568 |
| Sherlock p-value | 0.77 |
| Fetal beta | -0.403 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1257641 | chr14 | 99480394 | ROCK2 | 9475 | 0.14 | trans |
Section II. Transcriptome annotation
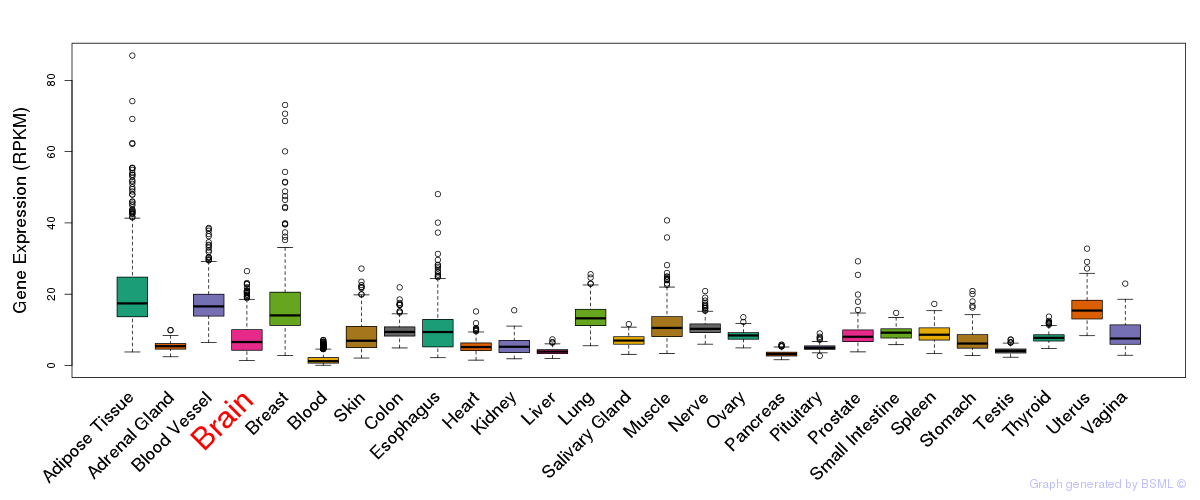
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DENND5A | 0.84 | 0.87 |
| LAMP1 | 0.84 | 0.82 |
| NPC1 | 0.83 | 0.81 |
| CNTN2 | 0.81 | 0.81 |
| JAM3 | 0.80 | 0.84 |
| ENDOD1 | 0.80 | 0.76 |
| USP54 | 0.80 | 0.80 |
| IPO13 | 0.79 | 0.77 |
| PTPDC1 | 0.79 | 0.79 |
| AP006216.1 | 0.79 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RP9P | -0.47 | -0.53 |
| ST20 | -0.44 | -0.47 |
| SPDYA | -0.43 | -0.46 |
| IL32 | -0.42 | -0.50 |
| SYCP3 | -0.42 | -0.44 |
| FAM159B | -0.42 | -0.55 |
| AC087071.1 | -0.40 | -0.36 |
| AL050337.1 | -0.39 | -0.37 |
| NANOS3 | -0.39 | -0.42 |
| NOX1 | -0.39 | -0.37 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR3 PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| SIG CHEMOTAXIS | 45 | 37 | All SZGR 2.0 genes in this pathway |
| SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | 35 | 25 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID PLK1 PATHWAY | 46 | 25 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR4 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D IN SEMAPHORIN SIGNALING | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | 27 | 21 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA COAMPLIFIED WITH MYCN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 20 HELA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |