Gene Page: CLOCK
Summary ?
| GeneID | 9575 |
| Symbol | CLOCK |
| Synonyms | KAT13D|bHLHe8 |
| Description | clock circadian regulator |
| Reference | MIM:601851|HGNC:HGNC:2082|Ensembl:ENSG00000134852|HPRD:03508|Vega:OTTHUMG00000102141 |
| Gene type | protein-coding |
| Map location | 4q12 |
| Pascal p-value | 0.05 |
| Sherlock p-value | 0.792 |
| Fetal beta | -0.55 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25530661 | 4 | 56301576 | CLOCK | 2.402E-4 | -0.316 | 0.037 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
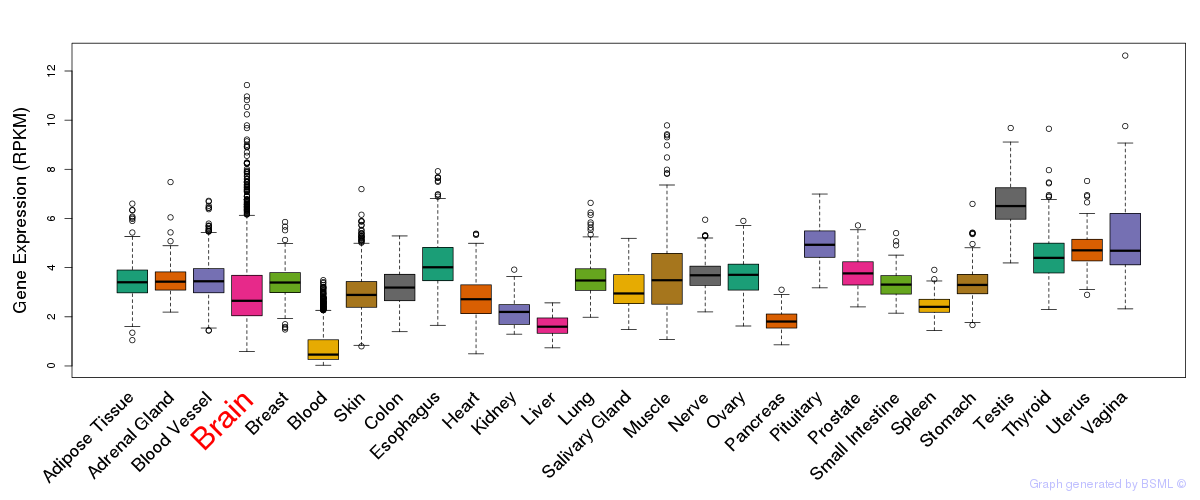
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRRT3 | 0.86 | 0.91 |
| NDRG4 | 0.86 | 0.87 |
| RPS6KA4 | 0.85 | 0.87 |
| PINK1 | 0.85 | 0.88 |
| COPS7A | 0.85 | 0.87 |
| PPAPDC3 | 0.84 | 0.87 |
| ANXA6 | 0.84 | 0.81 |
| GRIN1 | 0.84 | 0.86 |
| PLD3 | 0.84 | 0.86 |
| AP001107.1 | 0.83 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBMX2 | -0.50 | -0.53 |
| C9orf46 | -0.47 | -0.50 |
| EXOSC8 | -0.46 | -0.43 |
| GTF3C6 | -0.46 | -0.47 |
| C21orf57 | -0.45 | -0.46 |
| DYNLT1 | -0.44 | -0.51 |
| RPS25P8 | -0.44 | -0.53 |
| RPS8 | -0.44 | -0.46 |
| RPS20 | -0.44 | -0.54 |
| ZNF300 | -0.44 | -0.17 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 10198158 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004402 | histone acetyltransferase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008415 | acyltransferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007165 | signal transduction | TAS | 10198158 | |
| GO:0009648 | photoperiodism | TAS | 10198158 | |
| GO:0007623 | circadian rhythm | IEA | - | |
| GO:0007623 | circadian rhythm | TAS | 10198158 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IGI | 9576906 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005667 | transcription factor complex | IEA | - | |
| GO:0005667 | transcription factor complex | IPI | 9576906 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CIRCADIAN RHYTHM MAMMAL | 13 | 13 | All SZGR 2.0 genes in this pathway |
| PID CIRCADIAN PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | 23 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN CLOCK | 53 | 40 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| IVANOVSKA MIR106B TARGETS | 90 | 56 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 2306 | 2312 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-124/506 | 1205 | 1211 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-130/301 | 912 | 918 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-148/152 | 913 | 919 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-15/16/195/424/497 | 797 | 803 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 1069 | 1075 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-182 | 271 | 278 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-19 | 1048 | 1055 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-190 | 2118 | 2125 | 1A,m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-30-5p | 697 | 704 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-409-3p | 1078 | 1084 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-495 | 775 | 781 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-542-3p | 768 | 774 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-543 | 1023 | 1029 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCU | ||||
| miR-9 | 2054 | 2061 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 272 | 278 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.