Gene Page: PPM1F
Summary ?
| GeneID | 9647 |
| Symbol | PPM1F |
| Synonyms | CAMKP|CaMKPase|FEM-2|POPX2|hFEM-2 |
| Description | protein phosphatase, Mg2+/Mn2+ dependent 1F |
| Reference | HGNC:HGNC:19388|Ensembl:ENSG00000100034|HPRD:10163|Vega:OTTHUMG00000150835 |
| Gene type | protein-coding |
| Map location | 22q11.22 |
| Pascal p-value | 0.01 |
| Sherlock p-value | 0.768 |
| Fetal beta | 1.047 |
| eGene | Cerebellar Hemisphere Cerebellum Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17542473 | chr4 | 111182451 | PPM1F | 9647 | 0.08 | trans | ||
| rs56094396 | 22 | 22278727 | PPM1F | ENSG00000100034.9 | 2.128E-7 | 0.01 | 28482 | gtex_brain_putamen_basal |
| rs738858 | 22 | 22280917 | PPM1F | ENSG00000100034.9 | 5.633E-7 | 0.01 | 26292 | gtex_brain_putamen_basal |
| rs240064 | 22 | 22281638 | PPM1F | ENSG00000100034.9 | 1.277E-6 | 0.01 | 25571 | gtex_brain_putamen_basal |
| rs2329884 | 22 | 22282263 | PPM1F | ENSG00000100034.9 | 4.729E-7 | 0.01 | 24946 | gtex_brain_putamen_basal |
| rs894095 | 22 | 22282776 | PPM1F | ENSG00000100034.9 | 4.729E-7 | 0.01 | 24433 | gtex_brain_putamen_basal |
| . | 22 | 22283231 | PPM1F | ENSG00000100034.9 | 9.157E-7 | 0.01 | 23978 | gtex_brain_putamen_basal |
| rs33940969 | 22 | 22283232 | PPM1F | ENSG00000100034.9 | 9.684E-7 | 0.01 | 23977 | gtex_brain_putamen_basal |
| rs2083565 | 22 | 22284357 | PPM1F | ENSG00000100034.9 | 4.729E-7 | 0.01 | 22852 | gtex_brain_putamen_basal |
| rs5995257 | 22 | 22285987 | PPM1F | ENSG00000100034.9 | 4.409E-7 | 0.01 | 21222 | gtex_brain_putamen_basal |
| rs2027790 | 22 | 22287964 | PPM1F | ENSG00000100034.9 | 3.39E-7 | 0.01 | 19245 | gtex_brain_putamen_basal |
| rs94194 | 22 | 22288309 | PPM1F | ENSG00000100034.9 | 3.241E-6 | 0.01 | 18900 | gtex_brain_putamen_basal |
| rs5995268 | 22 | 22289031 | PPM1F | ENSG00000100034.9 | 5.041E-7 | 0.01 | 18178 | gtex_brain_putamen_basal |
| rs5995269 | 22 | 22289397 | PPM1F | ENSG00000100034.9 | 7.673E-7 | 0.01 | 17812 | gtex_brain_putamen_basal |
| rs240066 | 22 | 22292995 | PPM1F | ENSG00000100034.9 | 1.874E-6 | 0.01 | 14214 | gtex_brain_putamen_basal |
| rs5756181 | 22 | 22305008 | PPM1F | ENSG00000100034.9 | 2.034E-6 | 0.01 | 2201 | gtex_brain_putamen_basal |
| rs381849 | 22 | 22305713 | PPM1F | ENSG00000100034.9 | 2.513E-6 | 0.01 | 1496 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
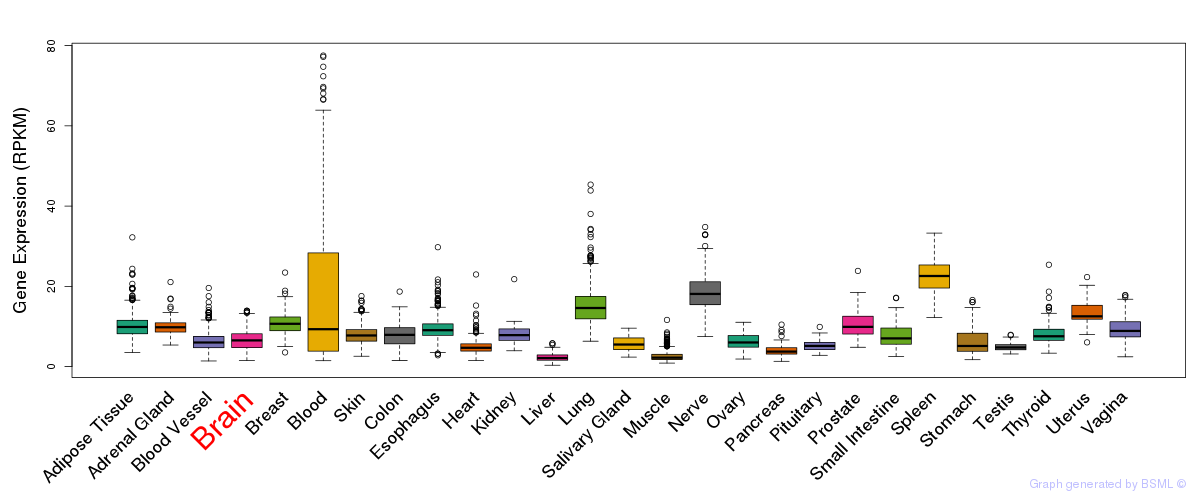
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF4B | 0.93 | 0.92 |
| IGBP1 | 0.92 | 0.88 |
| INTS7 | 0.91 | 0.92 |
| COMMD2 | 0.91 | 0.89 |
| ARG2 | 0.90 | 0.91 |
| KARS | 0.90 | 0.92 |
| CCT3 | 0.90 | 0.89 |
| POLR1E | 0.90 | 0.90 |
| EEF2 | 0.90 | 0.89 |
| TEX10 | 0.90 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.65 | -0.86 |
| AF347015.31 | -0.65 | -0.85 |
| AF347015.33 | -0.65 | -0.85 |
| FXYD1 | -0.64 | -0.87 |
| C5orf53 | -0.64 | -0.77 |
| HLA-F | -0.63 | -0.78 |
| AF347015.27 | -0.63 | -0.84 |
| IFI27 | -0.63 | -0.85 |
| HEPN1 | -0.63 | -0.77 |
| MT-CYB | -0.63 | -0.83 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004722 | protein serine/threonine phosphatase activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008287 | protein serine/threonine phosphatase complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN | 77 | 46 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| PATTERSON DOCETAXEL RESISTANCE | 29 | 20 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| GEORGANTAS HSC MARKERS | 71 | 47 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C CLUSTER UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C UP | 47 | 29 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO CISPLATIN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 942 | 948 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 941 | 948 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-149 | 1944 | 1950 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-200bc/429 | 719 | 725 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-217 | 864 | 870 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-299-5p | 3650 | 3656 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-342 | 2753 | 2759 | m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-377 | 2752 | 2758 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-96 | 750 | 756 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.