Gene Page: MRPL19
Summary ?
| GeneID | 9801 |
| Symbol | MRPL19 |
| Synonyms | L19mt|MRP-L15|MRP-L19|MRPL15|RLX1|RPML15 |
| Description | mitochondrial ribosomal protein L19 |
| Reference | MIM:611832|HGNC:HGNC:14052|HPRD:11370| |
| Gene type | protein-coding |
| Map location | 2p11.1-q11.2 |
| Pascal p-value | 0.663 |
| Sherlock p-value | 0.674 |
| Fetal beta | 0.332 |
| DMG | 2 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06647844 | 2 | 75888565 | MRPL19 | 1.59E-4 | 0.006 | 0.142 | DMG:Montano_2016 |
| cg02916181 | 2 | 75873822 | MRPL19 | 9.09E-9 | -0.018 | 4.12E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17689863 | chr2 | 75882398 | MRPL19 | 9801 | 0.02 | cis | ||
| rs17690224 | chr2 | 75892110 | MRPL19 | 9801 | 0.03 | cis | ||
| rs4853169 | chr2 | 75906971 | MRPL19 | 9801 | 0 | cis | ||
| rs4853169 | chr2 | 75906971 | MRPL19 | 9801 | 0.1 | trans | ||
| rs34827428 | 2 | 75876190 | MRPL19 | ENSG00000115364.9 | 7.644E-10 | 0 | 2281 | gtex_brain_ba24 |
| rs11688959 | 2 | 75878511 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 4602 | gtex_brain_ba24 |
| rs4853168 | 2 | 75879452 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 5543 | gtex_brain_ba24 |
| rs34356427 | 2 | 75881251 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 7342 | gtex_brain_ba24 |
| rs17689863 | 2 | 75882399 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 8490 | gtex_brain_ba24 |
| rs3732304 | 2 | 75882855 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 8946 | gtex_brain_ba24 |
| rs10165899 | 2 | 75883276 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 9367 | gtex_brain_ba24 |
| rs2190356 | 2 | 75885410 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 11501 | gtex_brain_ba24 |
| rs17690224 | 2 | 75892111 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 18202 | gtex_brain_ba24 |
| rs7573291 | 2 | 75893853 | MRPL19 | ENSG00000115364.9 | 2.54E-10 | 0 | 19944 | gtex_brain_ba24 |
| rs11692855 | 2 | 75896630 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 22721 | gtex_brain_ba24 |
| rs34645183 | 2 | 75897939 | MRPL19 | ENSG00000115364.9 | 4.18E-10 | 0 | 24030 | gtex_brain_ba24 |
| rs1986239 | 2 | 75900207 | MRPL19 | ENSG00000115364.9 | 1.484E-9 | 0 | 26298 | gtex_brain_ba24 |
| rs1986240 | 2 | 75900211 | MRPL19 | ENSG00000115364.9 | 7.907E-10 | 0 | 26302 | gtex_brain_ba24 |
| rs2422232 | 2 | 75900380 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 26471 | gtex_brain_ba24 |
| rs2051942 | 2 | 75901069 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 27160 | gtex_brain_ba24 |
| rs34081022 | 2 | 75902380 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 28471 | gtex_brain_ba24 |
| rs4853169 | 2 | 75906972 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 33063 | gtex_brain_ba24 |
| rs35450646 | 2 | 75911046 | MRPL19 | ENSG00000115364.9 | 4.246E-10 | 0 | 37137 | gtex_brain_ba24 |
| rs13391581 | 2 | 75913595 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 39686 | gtex_brain_ba24 |
| rs111252424 | 2 | 75914130 | MRPL19 | ENSG00000115364.9 | 2.815E-10 | 0 | 40221 | gtex_brain_ba24 |
| rs111525346 | 2 | 75914136 | MRPL19 | ENSG00000115364.9 | 2.815E-10 | 0 | 40227 | gtex_brain_ba24 |
| rs397841130 | 2 | 75915920 | MRPL19 | ENSG00000115364.9 | 2.298E-10 | 0 | 42011 | gtex_brain_ba24 |
| rs182810843 | 2 | 75919292 | MRPL19 | ENSG00000115364.9 | 3.047E-8 | 0 | 45383 | gtex_brain_ba24 |
| rs11677947 | 2 | 75927536 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 53627 | gtex_brain_ba24 |
| rs33915308 | 2 | 75928122 | MRPL19 | ENSG00000115364.9 | 2.505E-7 | 0 | 54213 | gtex_brain_ba24 |
| rs33971443 | 2 | 75931128 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 57219 | gtex_brain_ba24 |
| rs7583552 | 2 | 75932062 | MRPL19 | ENSG00000115364.9 | 4.394E-10 | 0 | 58153 | gtex_brain_ba24 |
| rs114255554 | 2 | 75932627 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 58718 | gtex_brain_ba24 |
| rs11681342 | 2 | 75933130 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 59221 | gtex_brain_ba24 |
| rs367550848 | 2 | 75934865 | MRPL19 | ENSG00000115364.9 | 1.125E-9 | 0 | 60956 | gtex_brain_ba24 |
| rs113730259 | 2 | 75939031 | MRPL19 | ENSG00000115364.9 | 4.195E-10 | 0 | 65122 | gtex_brain_ba24 |
| rs112846031 | 2 | 75939781 | MRPL19 | ENSG00000115364.9 | 4.188E-10 | 0 | 65872 | gtex_brain_ba24 |
| rs34240870 | 2 | 75942346 | MRPL19 | ENSG00000115364.9 | 3.162E-10 | 0 | 68437 | gtex_brain_ba24 |
| rs17741889 | 2 | 75942725 | MRPL19 | ENSG00000115364.9 | 3.043E-10 | 0 | 68816 | gtex_brain_ba24 |
| rs11680870 | 2 | 75850411 | MRPL19 | ENSG00000115364.9 | 1.393E-7 | 0 | -23498 | gtex_brain_putamen_basal |
| rs67929017 | 2 | 75853353 | MRPL19 | ENSG00000115364.9 | 2.838E-8 | 0 | -20556 | gtex_brain_putamen_basal |
| rs188346656 | 2 | 75854670 | MRPL19 | ENSG00000115364.9 | 2.198E-6 | 0 | -19239 | gtex_brain_putamen_basal |
| rs4853165 | 2 | 75855463 | MRPL19 | ENSG00000115364.9 | 4.059E-8 | 0 | -18446 | gtex_brain_putamen_basal |
| rs6747017 | 2 | 75857547 | MRPL19 | ENSG00000115364.9 | 5.367E-9 | 0 | -16362 | gtex_brain_putamen_basal |
| rs35908333 | 2 | 75862620 | MRPL19 | ENSG00000115364.9 | 3.32E-9 | 0 | -11289 | gtex_brain_putamen_basal |
| rs1859710 | 2 | 75862932 | MRPL19 | ENSG00000115364.9 | 3.32E-9 | 0 | -10977 | gtex_brain_putamen_basal |
| rs6711562 | 2 | 75864327 | MRPL19 | ENSG00000115364.9 | 1.612E-9 | 0 | -9582 | gtex_brain_putamen_basal |
| rs6754000 | 2 | 75864616 | MRPL19 | ENSG00000115364.9 | 7.643E-10 | 0 | -9293 | gtex_brain_putamen_basal |
| rs10570064 | 2 | 75865935 | MRPL19 | ENSG00000115364.9 | 7.729E-10 | 0 | -7974 | gtex_brain_putamen_basal |
| rs36057194 | 2 | 75872399 | MRPL19 | ENSG00000115364.9 | 7.987E-10 | 0 | -1510 | gtex_brain_putamen_basal |
| rs2286239 | 2 | 75874390 | MRPL19 | ENSG00000115364.9 | 1.919E-9 | 0 | 481 | gtex_brain_putamen_basal |
| rs34827428 | 2 | 75876190 | MRPL19 | ENSG00000115364.9 | 8.11E-14 | 0 | 2281 | gtex_brain_putamen_basal |
| rs11688959 | 2 | 75878511 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 4602 | gtex_brain_putamen_basal |
| rs4853168 | 2 | 75879452 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 5543 | gtex_brain_putamen_basal |
| rs34356427 | 2 | 75881251 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 7342 | gtex_brain_putamen_basal |
| rs17689863 | 2 | 75882399 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 8490 | gtex_brain_putamen_basal |
| rs3732304 | 2 | 75882855 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 8946 | gtex_brain_putamen_basal |
| rs10165899 | 2 | 75883276 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 9367 | gtex_brain_putamen_basal |
| rs2190356 | 2 | 75885410 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 11501 | gtex_brain_putamen_basal |
| rs17690224 | 2 | 75892111 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 18202 | gtex_brain_putamen_basal |
| rs7573291 | 2 | 75893853 | MRPL19 | ENSG00000115364.9 | 1.172E-13 | 0 | 19944 | gtex_brain_putamen_basal |
| rs11692855 | 2 | 75896630 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 22721 | gtex_brain_putamen_basal |
| rs34645183 | 2 | 75897939 | MRPL19 | ENSG00000115364.9 | 1.687E-13 | 0 | 24030 | gtex_brain_putamen_basal |
| rs1986239 | 2 | 75900207 | MRPL19 | ENSG00000115364.9 | 8.862E-12 | 0 | 26298 | gtex_brain_putamen_basal |
| rs1986240 | 2 | 75900211 | MRPL19 | ENSG00000115364.9 | 1.971E-12 | 0 | 26302 | gtex_brain_putamen_basal |
| rs2422232 | 2 | 75900380 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 26471 | gtex_brain_putamen_basal |
| rs2051942 | 2 | 75901069 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 27160 | gtex_brain_putamen_basal |
| rs34081022 | 2 | 75902380 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 28471 | gtex_brain_putamen_basal |
| rs4853169 | 2 | 75906972 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 33063 | gtex_brain_putamen_basal |
| rs35450646 | 2 | 75911046 | MRPL19 | ENSG00000115364.9 | 1.697E-13 | 0 | 37137 | gtex_brain_putamen_basal |
| rs13391581 | 2 | 75913595 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 39686 | gtex_brain_putamen_basal |
| rs111252424 | 2 | 75914130 | MRPL19 | ENSG00000115364.9 | 1.589E-13 | 0 | 40221 | gtex_brain_putamen_basal |
| rs111525346 | 2 | 75914136 | MRPL19 | ENSG00000115364.9 | 1.589E-13 | 0 | 40227 | gtex_brain_putamen_basal |
| rs397841130 | 2 | 75915920 | MRPL19 | ENSG00000115364.9 | 1.724E-13 | 0 | 42011 | gtex_brain_putamen_basal |
| rs182810843 | 2 | 75919292 | MRPL19 | ENSG00000115364.9 | 7.504E-13 | 0 | 45383 | gtex_brain_putamen_basal |
| rs11677947 | 2 | 75927536 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 53627 | gtex_brain_putamen_basal |
| rs33915308 | 2 | 75928122 | MRPL19 | ENSG00000115364.9 | 4.604E-12 | 0 | 54213 | gtex_brain_putamen_basal |
| rs33971443 | 2 | 75931128 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 57219 | gtex_brain_putamen_basal |
| rs7583552 | 2 | 75932062 | MRPL19 | ENSG00000115364.9 | 1.723E-13 | 0 | 58153 | gtex_brain_putamen_basal |
| rs114255554 | 2 | 75932627 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 58718 | gtex_brain_putamen_basal |
| rs11681342 | 2 | 75933130 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 59221 | gtex_brain_putamen_basal |
| rs367550848 | 2 | 75934865 | MRPL19 | ENSG00000115364.9 | 3.332E-12 | 0 | 60956 | gtex_brain_putamen_basal |
| rs113730259 | 2 | 75939031 | MRPL19 | ENSG00000115364.9 | 1.688E-13 | 0 | 65122 | gtex_brain_putamen_basal |
| rs112846031 | 2 | 75939781 | MRPL19 | ENSG00000115364.9 | 1.689E-13 | 0 | 65872 | gtex_brain_putamen_basal |
| rs34240870 | 2 | 75942346 | MRPL19 | ENSG00000115364.9 | 1.983E-13 | 0 | 68437 | gtex_brain_putamen_basal |
| rs17741889 | 2 | 75942725 | MRPL19 | ENSG00000115364.9 | 2.225E-13 | 0 | 68816 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
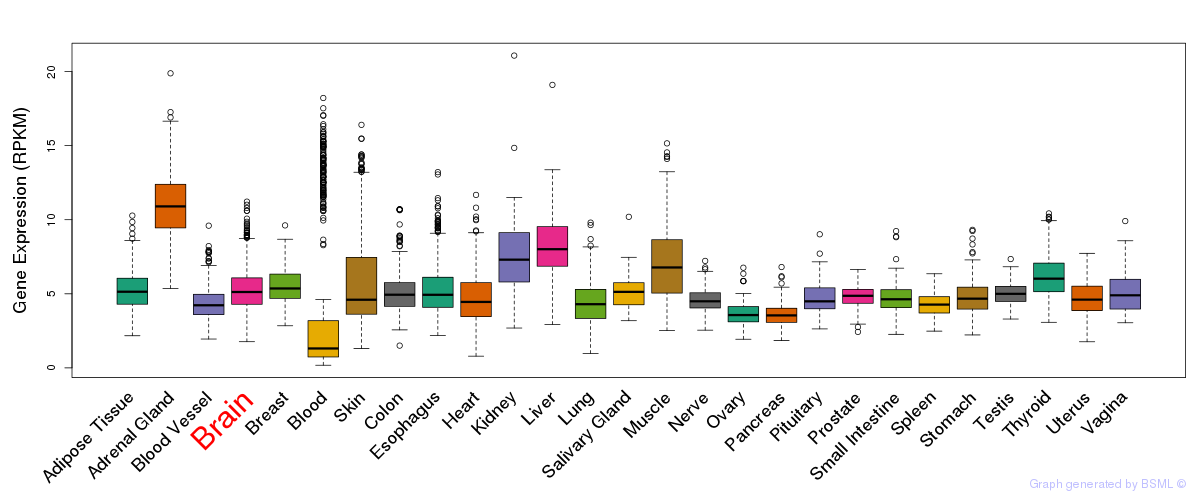
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC11A2 | 0.75 | 0.74 |
| ALG13 | 0.73 | 0.74 |
| MAP3K12 | 0.73 | 0.74 |
| COG7 | 0.72 | 0.74 |
| MUM1 | 0.71 | 0.72 |
| RP11-791G16.2 | 0.71 | 0.79 |
| NDST2 | 0.71 | 0.72 |
| NSUN2 | 0.71 | 0.72 |
| MAT2A | 0.71 | 0.77 |
| HMGXB3 | 0.70 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.64 | -0.61 |
| MT-CO2 | -0.60 | -0.56 |
| AF347015.31 | -0.58 | -0.55 |
| AF347015.27 | -0.57 | -0.53 |
| AF347015.8 | -0.56 | -0.54 |
| AC098691.2 | -0.55 | -0.57 |
| MT-CYB | -0.53 | -0.50 |
| AF347015.33 | -0.53 | -0.48 |
| C1orf54 | -0.52 | -0.56 |
| HIGD1B | -0.51 | -0.49 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE DN | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR | 74 | 47 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |