Gene Page: CDK4
Summary ?
| GeneID | 1019 |
| Symbol | CDK4 |
| Synonyms | CMM3|PSK-J3 |
| Description | cyclin-dependent kinase 4 |
| Reference | MIM:123829|HGNC:HGNC:1773|Ensembl:ENSG00000135446|HPRD:00447|Vega:OTTHUMG00000170382 |
| Gene type | protein-coding |
| Map location | 12q14 |
| Pascal p-value | 0.601 |
| Sherlock p-value | 0.833 |
| Fetal beta | 1.769 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1875 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22715954 | 12 | 58146426 | CDK4 | 9.25E-9 | -0.022 | 4.15E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
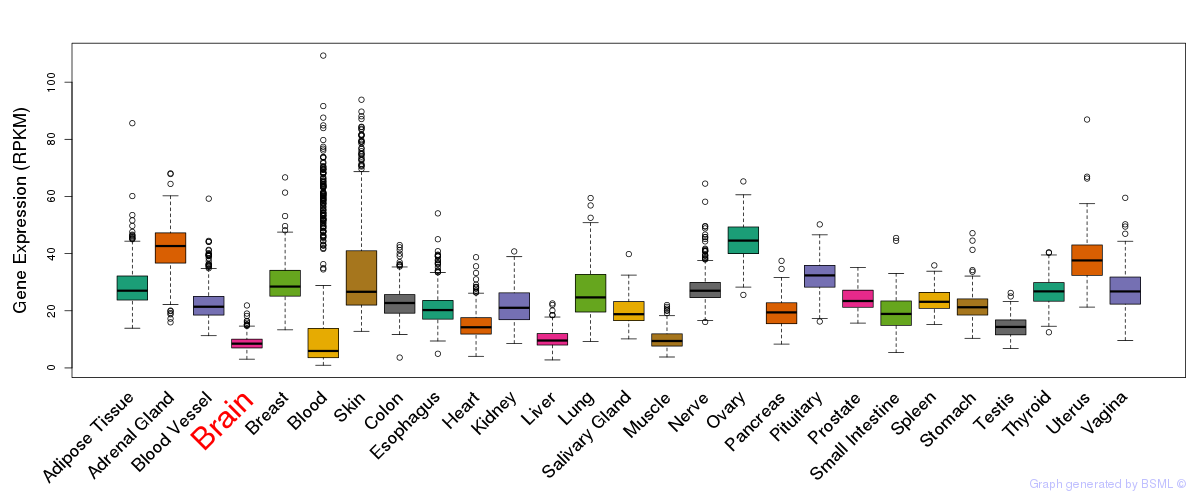
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TUBGCP6 | 0.92 | 0.94 |
| WDR59 | 0.90 | 0.91 |
| RHOT2 | 0.89 | 0.88 |
| LZTR1 | 0.89 | 0.91 |
| SH2B1 | 0.88 | 0.91 |
| NDST2 | 0.88 | 0.90 |
| COG7 | 0.88 | 0.90 |
| ZSWIM1 | 0.88 | 0.89 |
| ANKS3 | 0.88 | 0.90 |
| ABCC10 | 0.87 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.72 | -0.78 |
| AF347015.31 | -0.71 | -0.73 |
| AF347015.27 | -0.70 | -0.72 |
| MT-CO2 | -0.69 | -0.71 |
| AF347015.8 | -0.68 | -0.70 |
| MT-CYB | -0.67 | -0.68 |
| AF347015.33 | -0.65 | -0.66 |
| AF347015.15 | -0.64 | -0.66 |
| MT-ATP8 | -0.62 | -0.73 |
| AF347015.2 | -0.62 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004693 | cyclin-dependent protein kinase activity | IEA | neuron (GO term level: 8) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9106657 |11896535 |15232106 |15558030 |16169070 |17274640 |17353931 |17909018 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004672 | protein kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000082 | G1/S transition of mitotic cell cycle | IMP | 7603984 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0010468 | regulation of gene expression | IMP | 17420273 | |
| GO:0051301 | cell division | IEA | - | |
| GO:0048146 | positive regulation of fibroblast proliferation | IMP | 17420273 | |
| GO:0051726 | regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | IEA | - | |
| GO:0005829 | cytosol | EXP | 9106657 | |
| GO:0005634 | nucleus | IDA | 16109376 | |
| GO:0005654 | nucleoplasm | EXP | 9190208 |16782892 | |
| GO:0005667 | transcription factor complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTR3B | ARP11 | ARP3BETA | DKFZp686O24114 | ARP3 actin-related protein 3 homolog B (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| BAT3 | BAG-6 | BAG6 | D6S52E | G3 | HLA-B associated transcript 3 | Two-hybrid | BioGRID | 16169070 |
| BIRC5 | API4 | EPR-1 | baculoviral IAP repeat-containing 5 | - | HPRD,BioGRID | 10713676 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 9244350 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 associates with CDK4 | BIND | 9244350 |
| CAPNS1 | 30K | CALPAIN4 | CANP | CANPS | CAPN4 | CDPS | calpain, small subunit 1 | Two-hybrid | BioGRID | 16169070 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD | 9150368 |9837900 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | CDK4 interacts with Cyclin D1. This interaction was modelled on a demonstrated interaction between human CDK4 and Cyclin D1 from an unspecified species. | BIND | 9228064 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | CCND1 (cyclin D1) interacts with CDK4. | BIND | 8662825 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8259215 |9837900 |10580009 |11360184 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | CCND2 interacts with CDK4 | BIND | 11291051 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | - | HPRD,BioGRID | 11358847 |
| CCND3 | - | cyclin D3 | - | HPRD,BioGRID | 10342870 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | - | HPRD,BioGRID | 8666233 |8703009 |
| CDC7 | CDC7L1 | HsCDC7 | Hsk1 | MGC117361 | MGC126237 | MGC126238 | huCDC7 | cell division cycle 7 homolog (S. cerevisiae) | - | HPRD | 15232106 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Cdk4 interacts with p21. | BIND | 8999999 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | - | HPRD,BioGRID | 10908655 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | p16 interacts with cdk4. This interaction was modeled on a demonstrated interaction between human p16 and cdk4 from an unspecified species. | BIND | 9482104 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | CDKN2A interacts with CDK4 | BIND | 11556834 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | - | HPRD,BioGRID | 9228064 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | - | HPRD | 9228064 |15232106 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | p16 interacts with CDK4. This interaction was modeled on a demonstrated interaction between human p16 and CDK4 from an unspecified species. | BIND | 10491434 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | p16 interacts with cdk4. | BIND | 8805225 |
| CDKN2B | CDK4I | INK4B | MTS2 | P15 | TP15 | p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | - | HPRD | 7478582 |9111314 |9163429 |
| CDKN2B | CDK4I | INK4B | MTS2 | P15 | TP15 | p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | in vitro in vivo Two-hybrid | BioGRID | 9163429 |16169070 |16189514 |
| CDKN2C | INK4C | p18 | p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | - | HPRD,BioGRID | 8001816 |
| CDKN2C | INK4C | p18 | p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | An unspecified isoform of p18 interacts with cdk4. This interaction was modeled on a demonstrated interaction between human p18 and cdk4 from an unspecified species. | BIND | 9482104 |
| CDKN2D | INK4D | p19 | p19-INK4D | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | Two-hybrid | BioGRID | 16189514 |
| CEBPA | C/EBP-alpha | CEBP | CCAAT/enhancer binding protein (C/EBP), alpha | - | HPRD,BioGRID | 11684017 |
| CNOT7 | CAF1 | hCAF-1 | CCR4-NOT transcription complex, subunit 7 | - | HPRD,BioGRID | 10602502 |
| DBNL | ABP1 | HIP-55 | SH3P7 | drebrin-like | Affinity Capture-MS | BioGRID | 17353931 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | Affinity Capture-MS | BioGRID | 17353931 |
| IFI27 | FAM14D | ISG12 | ISG12A | P27 | interferon, alpha-inducible protein 27 | - | HPRD | 9837900 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | Affinity Capture-MS | BioGRID | 17353931 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | MYC (c-Myc) interacts with CDK4 promoter. | BIND | 12857727 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD,BioGRID | 10022835 |10601020 |
| ORC3L | LAT | LATHEO | ORC3 | origin recognition complex, subunit 3-like (yeast) | - | HPRD | 15232106 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | Affinity Capture-Western | BioGRID | 8259215 |
| PSMD10 | dJ889N15.2 | p28 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | - | HPRD,BioGRID | 11779854 |
| PTMA | MGC104802 | TMSA | prothymosin, alpha | - | HPRD | 11310559 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | CDK4 phosphorylates Rb carboxy-terminus. This interaction was modelled on a demonstrated interaction between human Rb and mouse CDK4. | BIND | 15241418 |
| RFC1 | A1 | MGC51786 | MHCBFB | PO-GA | RECC1 | RFC | RFC140 | replication factor C (activator 1) 1, 145kDa | - | HPRD,BioGRID | 10353443 |
| RPRM | FLJ90327 | REPRIMO | reprimo, TP53 dependent G2 arrest mediator candidate | Affinity Capture-MS | BioGRID | 17353931 |
| SERTAD1 | SEI1 | TRIP-Br1 | SERTA domain containing 1 | - | HPRD,BioGRID | 10580009 |15065884 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | Two-hybrid | BioGRID | 16169070 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | CDK4 phosphorylates Smad2. This interaction was modelled on a demonstrated interaction between Smad2 from an unspecified species and mouse CDK4. | BIND | 15241418 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | CDK4 phosphorylates Smad3 on Thr8, Thr179, Ser204, Ser208 and Ser213. This interaction was modelled on a demonstrated interaction between Smad3 from an unspecified species and mouse CDK4. | BIND | 15241418 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | Affinity Capture-MS | BioGRID | 17353931 |
| SMC3 | BAM | BMH | CDLS3 | CSPG6 | HCAP | SMC3L1 | structural maintenance of chromosomes 3 | Affinity Capture-MS | BioGRID | 17353931 |
| UBXN1 | 2B28 | UBXD10 | UBX domain protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| VTA1 | C6orf55 | DRG-1 | DRG1 | FLJ27228 | HSPC228 | LIP5 | My012 | SBP1 | Vps20-associated 1 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELLCYCLE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53 PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RB PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| SA G1 AND S PHASES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SA REG CASCADE OF CYCLIN EXPR | 13 | 7 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC UP | 23 | 17 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB UP | 29 | 20 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 1 | 45 | 27 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 21 | 9 | 5 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLIFIED IN SOFT TISSUE CANCER | 13 | 7 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 5 | 18 | 11 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 12Q13 Q21 AMPLICON | 46 | 21 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER UP | 75 | 36 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| SCHUHMACHER MYC TARGETS UP | 80 | 57 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC | 50 | 32 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 3 UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD DN | 56 | 35 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| MATZUK CENTRAL FOR FEMALE FERTILITY | 29 | 25 | All SZGR 2.0 genes in this pathway |
| MATZUK LUTEAL GENES | 8 | 5 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| ZHAN VARIABLE EARLY DIFFERENTIATION GENES DN | 30 | 19 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LE NEURONAL DIFFERENTIATION DN | 19 | 16 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 131 | 138 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-486 | 258 | 264 | 1A | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.