Gene Page: RBPMS
Summary ?
| GeneID | 11030 |
| Symbol | RBPMS |
| Synonyms | HERMES |
| Description | RNA binding protein with multiple splicing |
| Reference | MIM:601558|HGNC:HGNC:19097|Ensembl:ENSG00000157110|HPRD:11870|Vega:OTTHUMG00000163845 |
| Gene type | protein-coding |
| Map location | 8p12 |
| Pascal p-value | 0.66 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.039 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs2979481 | 8 | 0 | null | 1.493 | RBPMS | null |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27258878 | 8 | 30407398 | RBPMS | 1.69E-5 | 0.255 | 0.015 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | RBPMS | 11030 | 0.18 | trans |
Section II. Transcriptome annotation
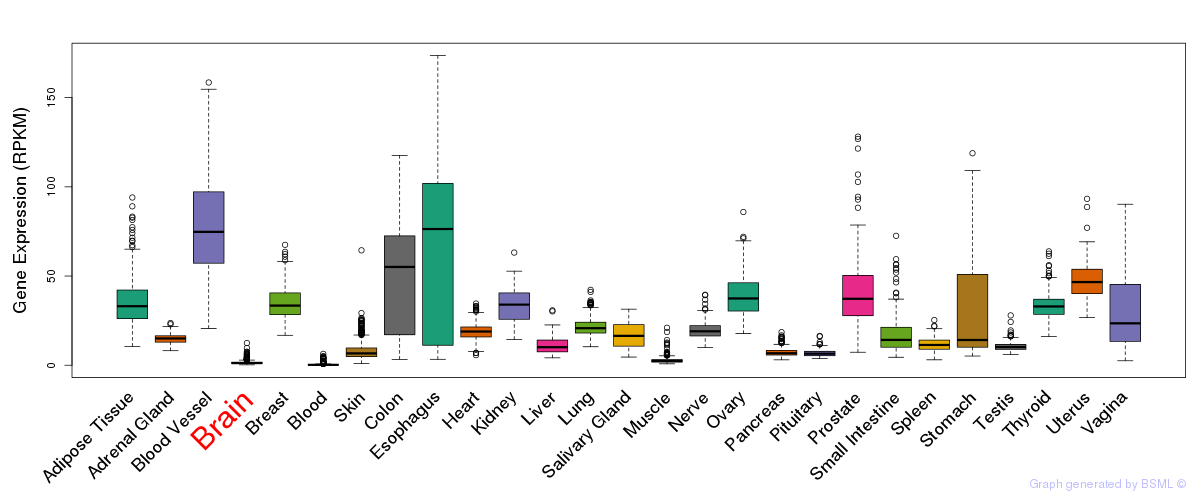
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBM19 | 0.90 | 0.90 |
| C17orf69 | 0.89 | 0.89 |
| SRCAP | 0.89 | 0.88 |
| TNRC6A | 0.89 | 0.90 |
| SETD1A | 0.89 | 0.88 |
| BAT2 | 0.89 | 0.88 |
| GIGYF1 | 0.88 | 0.91 |
| TCOF1 | 0.88 | 0.89 |
| RBM28 | 0.88 | 0.87 |
| PCNT | 0.88 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.66 | -0.70 |
| AF347015.21 | -0.64 | -0.77 |
| B2M | -0.64 | -0.66 |
| AF347015.27 | -0.64 | -0.68 |
| IFI27 | -0.63 | -0.66 |
| C1orf54 | -0.63 | -0.76 |
| SRGN | -0.62 | -0.68 |
| MT-CO2 | -0.62 | -0.67 |
| VAMP5 | -0.62 | -0.71 |
| HIGD1B | -0.62 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003723 | RNA binding | TAS | 8855282 | |
| GO:0005515 | protein binding | IPI | 16189514 |16713569 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006396 | RNA processing | TAS | 8855282 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGAP9 | 10C | FLJ16525 | MGC1295 | RGL1 | Rho GTPase activating protein 9 | Two-hybrid | BioGRID | 16189514 |
| C12orf12 | MGC26598 | chromosome 12 open reading frame 12 | Two-hybrid | BioGRID | 16189514 |
| C16orf48 | DAKV6410 | DKFZp434A1319 | chromosome 16 open reading frame 48 | Two-hybrid | BioGRID | 16189514 |
| C1orf94 | MGC15882 | chromosome 1 open reading frame 94 | Two-hybrid | BioGRID | 16189514 |
| C2orf51 | FLJ25369 | TSC21 | chromosome 2 open reading frame 51 | Two-hybrid | BioGRID | 16189514 |
| C9orf100 | FLJ14642 | MGC44886 | RP11-331F9.7 | chromosome 9 open reading frame 100 | Two-hybrid | BioGRID | 16189514 |
| C9orf86 | FLJ10101 | FLJ13045 | Parf | RBEL1 | bA216L13.9 | pp8875 | chromosome 9 open reading frame 86 | Two-hybrid | BioGRID | 16189514 |
| CCNK | CPR4 | MGC9113 | cyclin K | Two-hybrid | BioGRID | 16189514 |
| CDC23 | APC8 | cell division cycle 23 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| CDC42EP1 | BORG5 | CEP1 | MGC15316 | MSE55 | CDC42 effector protein (Rho GTPase binding) 1 | Two-hybrid | BioGRID | 16189514 |
| CNNM3 | ACDP3 | DKFZp434I1016 | FLJ20018 | cyclin M3 | Two-hybrid | BioGRID | 16189514 |
| DAZAP2 | KIAA0058 | MGC14319 | MGC766 | PRTB | DAZ associated protein 2 | Two-hybrid | BioGRID | 16189514 |
| DTX2 | KIAA1528 | MGC71098 | RNF58 | deltex homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Two-hybrid | BioGRID | 16189514 |
| EYA2 | EAB1 | MGC10614 | eyes absent homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| FAM103A1 | C15orf18 | HsT19360 | MGC102778 | MGC2560 | family with sequence similarity 103, member A1 | Two-hybrid | BioGRID | 16189514 |
| FASTK | FAST | Fas-activated serine/threonine kinase | Two-hybrid | BioGRID | 16189514 |
| FOXS1 | FKHL18 | FREAC10 | MGC4544 | forkhead box S1 | Two-hybrid | BioGRID | 16189514 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| GRAP2 | GADS | GRAP-2 | GRB2L | GRBLG | GRID | GRPL | GrbX | Grf40 | Mona | P38 | GRB2-related adaptor protein 2 | Two-hybrid | BioGRID | 16189514 |
| HEYL | HRT3 | MGC12623 | bHLHb33 | hairy/enhancer-of-split related with YRPW motif-like | Two-hybrid | BioGRID | 16189514 |
| HOXA9 | ABD-B | HOX1 | HOX1.7 | HOX1G | MGC1934 | homeobox A9 | Two-hybrid | BioGRID | 16189514 |
| HOXB9 | HOX-2.5 | HOX2 | HOX2E | homeobox B9 | Two-hybrid | BioGRID | 16189514 |
| KIAA0182 | GSE1 | KIAA0182 | Two-hybrid | BioGRID | 16189514 |
| LRRC41 | MGC126571 | MGC126573 | MUF1 | PP7759 | RP4-636H5.2 | leucine rich repeat containing 41 | Two-hybrid | BioGRID | 16189514 |
| LZTS2 | KIAA1813 | LAPSER1 | leucine zipper, putative tumor suppressor 2 | Two-hybrid | BioGRID | 16189514 |
| NEU4 | MGC102757 | MGC18222 | sialidase 4 | Two-hybrid | BioGRID | 16189514 |
| OTX1 | FLJ38361 | MGC15736 | orthodenticle homeobox 1 | Two-hybrid | BioGRID | 16189514 |
| PDLIM4 | RIL | PDZ and LIM domain 4 | Two-hybrid | BioGRID | 16189514 |
| PHF1 | MTF2L2 | PCL1 | PHF2 | PHD finger protein 1 | Two-hybrid | BioGRID | 16189514 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Two-hybrid | BioGRID | 16189514 |
| PITX1 | BFT | POTX | PTX1 | paired-like homeodomain 1 | Two-hybrid | BioGRID | 16189514 |
| POM121 | DKFZp586G1822 | DKFZp586P2220 | FLJ41820 | KIAA0618 | MGC3792 | POM121A | POM121 membrane glycoprotein (rat) | Two-hybrid | BioGRID | 16189514 |
| RBM9 | FOX2 | Fox-2 | HNRBP2 | HRNBP2 | RTA | dJ106I20.3 | fxh | RNA binding motif protein 9 | Two-hybrid | BioGRID | 16189514 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| RHOXF2 | PEPP-2 | PEPP2 | THG1 | Rhox homeobox family, member 2 | Two-hybrid | BioGRID | 16189514 |
| SERF2 | 4F5REL | FAM2C | FLJ20431 | FLJ37527 | FLJ38557 | H4F5rel | HsT17089 | MGC48826 | small EDRK-rich factor 2 | Two-hybrid | BioGRID | 16189514 |
| SF1 | D11S636 | ZFM1 | ZNF162 | splicing factor 1 | Two-hybrid | BioGRID | 16189514 |
| SMUG1 | FDG | HMUDG | MGC104370 | UNG3 | single-strand-selective monofunctional uracil-DNA glycosylase 1 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| STRBP | DKFZp434N214 | FLJ11307 | FLJ14223 | FLJ14984 | ILF3L | MGC21529 | MGC3405 | SPNR | p74 | spermatid perinuclear RNA binding protein | Two-hybrid | BioGRID | 16189514 |
| TOLLIP | FLJ33531 | IL-1RAcPIP | toll interacting protein | Two-hybrid | BioGRID | 16189514 |
| TRIP13 | 16E1BP | thyroid hormone receptor interactor 13 | Two-hybrid | BioGRID | 16189514 |
| ZC3H10 | FLJ14451 | ZC3HDC10 | zinc finger CCCH-type containing 10 | Two-hybrid | BioGRID | 16189514 |
| ZNF581 | FLJ22550 | HSPC189 | zinc finger protein 581 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN | 47 | 32 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| CASTELLANO HRAS AND NRAS TARGETS DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS DN | 14 | 14 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD DN | 36 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE NORMAL DN | 33 | 27 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER DN | 40 | 33 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS DN | 57 | 42 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| GEORGANTAS HSC MARKERS | 71 | 47 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS UP | 45 | 28 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL UP | 51 | 32 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 10 | 33 | 22 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH EVI1 | 25 | 15 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST VS SYNOVIAL SARCOMA DN | 20 | 17 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| GENTLES LEUKEMIC STEM CELL UP | 29 | 15 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-145 | 303 | 309 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-329 | 220 | 226 | m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-377 | 222 | 228 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-485-5p | 364 | 370 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.