Gene Page: DSP
Summary ?
| GeneID | 1832 |
| Symbol | DSP |
| Synonyms | DCWHKTA|DP |
| Description | desmoplakin |
| Reference | MIM:125647|HGNC:HGNC:3052|Ensembl:ENSG00000096696|HPRD:00513|Vega:OTTHUMG00000014212 |
| Gene type | protein-coding |
| Map location | 6p24 |
| Pascal p-value | 0.788 |
| Fetal beta | 0.343 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | G2Cdb.humanARC G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21068339 | 6 | 7542819 | DSP | 3.77E-10 | -0.011 | 7.51E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
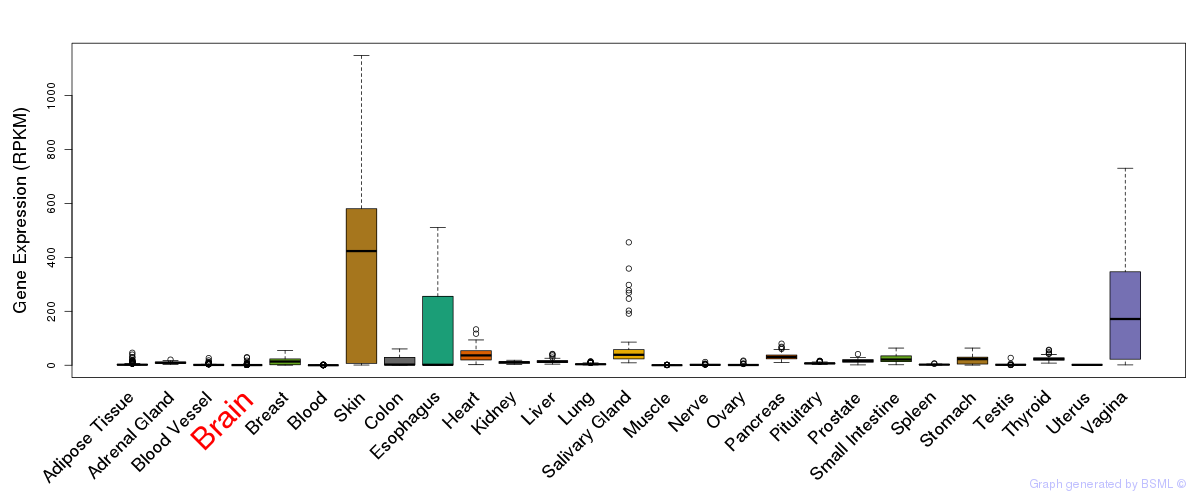
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PEF1 | 0.80 | 0.81 |
| STOML1 | 0.77 | 0.81 |
| ABHD14A | 0.77 | 0.80 |
| SH3GLB2 | 0.76 | 0.83 |
| ABHD8 | 0.76 | 0.79 |
| MTCH1 | 0.76 | 0.81 |
| RPUSD3 | 0.76 | 0.79 |
| C6orf154 | 0.75 | 0.79 |
| DNAJC30 | 0.75 | 0.78 |
| SH3BP1 | 0.75 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NSBP1 | -0.52 | -0.59 |
| EIF5B | -0.51 | -0.56 |
| AF347015.18 | -0.43 | -0.34 |
| ANP32B | -0.42 | -0.49 |
| RAB13 | -0.41 | -0.49 |
| AC010300.1 | -0.41 | -0.48 |
| AC005921.3 | -0.39 | -0.55 |
| GPR125 | -0.38 | -0.36 |
| CWF19L2 | -0.38 | -0.43 |
| ANP32C | -0.37 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005200 | structural constituent of cytoskeleton | TAS | 9887343 | |
| GO:0030674 | protein binding, bridging | IDA | 10908733 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008544 | epidermis development | TAS | 9887343 | |
| GO:0018149 | peptide cross-linking | IDA | 10908733 | |
| GO:0030216 | keratinocyte differentiation | IDA | 10908733 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001533 | cornified envelope | IDA | 10908733 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARID3A | BRIGHT | DRIL1 | DRIL3 | E2FBP1 | AT rich interactive domain 3A (BRIGHT-like) | - | HPRD | 9780002 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | - | HPRD | 15500642 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | - | HPRD | 15500642 |
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | - | HPRD | 15500642 |
| CASP6 | MCH2 | caspase 6, apoptosis-related cysteine peptidase | - | HPRD | 15500642 |
| CASP7 | CMH-1 | ICE-LAP3 | MCH3 | caspase 7, apoptosis-related cysteine peptidase | - | HPRD | 15500642 |
| CASP9 | APAF-3 | APAF3 | CASPASE-9c | ICE-LAP6 | MCH6 | caspase 9, apoptosis-related cysteine peptidase | - | HPRD | 15500642 |
| CDH5 | 7B4 | CD144 | FLJ17376 | cadherin 5, type 2 (vascular endothelium) | - | HPRD,BioGRID | 9739078 |
| CSTA | STF1 | STFA | cystatin A (stefin A) | Reconstituted Complex | BioGRID | 8999895 |
| DES | CMD1I | CSM1 | CSM2 | FLJ12025 | FLJ39719 | FLJ41013 | FLJ41793 | desmin | - | HPRD,BioGRID | 9261168 |
| DSC1 | CDHF1 | DG2/DG3 | desmocollin 1 | - | HPRD,BioGRID | 9606214 |
| DSP | DPI | DPII | desmoplakin | - | HPRD,BioGRID | 12101406 |
| EVPL | EVPK | envoplakin | - | HPRD | 8999895 |
| IKBKE | IKK-i | IKKE | IKKI | KIAA0151 | MGC125294 | MGC125295 | MGC125297 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon | - | HPRD | 14743216 |
| IVL | - | involucrin | Reconstituted Complex | BioGRID | 8999895 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | - | HPRD,BioGRID | 9348293 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | Plakoglobin interacts with desmoplakin. | BIND | 12426320 |
| KRT1 | CK1 | EHK1 | K1 | KRT1A | keratin 1 | - | HPRD,BioGRID | 9261168 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD | 14743216 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | - | HPRD | 14743216 |
| NFKBIB | IKBB | TRIP9 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | - | HPRD | 14743216 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | - | HPRD,BioGRID | 10801826 |
| PKP1 | B6P | MGC138829 | plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) | - | HPRD,BioGRID | 10852826 |
| PKP2 | ARVD9 | plakophilin 2 | - | HPRD,BioGRID | 11790773 |
| PKP3 | - | plakophilin 3 | PKP3 interacts with DSP(DP) | BIND | 12707304 |
| PKP3 | - | plakophilin 3 | Two-hybrid | BioGRID | 12707304 |
| PKP4 | FLJ31261 | FLJ42243 | p0071 | plakophilin 4 | p0071 interacts with desmoplakin. | BIND | 12426320 |
| PKP4 | FLJ31261 | FLJ42243 | p0071 | plakophilin 4 | Two-hybrid | BioGRID | 12615965 |
| RELB | I-REL | IREL | v-rel reticuloendotheliosis viral oncogene homolog B | - | HPRD | 14743216 |
| STK24 | MST-3 | MST3 | MST3B | STE20 | STK3 | serine/threonine kinase 24 (STE20 homolog, yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| VIM | FLJ36605 | vimentin | - | HPRD,BioGRID | 9261168 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | 12 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| MIDORIKAWA AMPLIFIED IN LIVER CANCER | 55 | 38 | All SZGR 2.0 genes in this pathway |
| AIGNER ZEB1 TARGETS | 35 | 15 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS UP | 28 | 16 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 24HR | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 6 | 75 | 48 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C6 | 5 | 5 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |