Gene Page: EPHB2
Summary ?
| GeneID | 2048 |
| Symbol | EPHB2 |
| Synonyms | CAPB|DRT|EK5|EPHT3|ERK|Hek5|PCBC|Tyro5 |
| Description | EPH receptor B2 |
| Reference | MIM:600997|HGNC:HGNC:3393|HPRD:02997| |
| Gene type | protein-coding |
| Map location | 1p36.1-p35 |
| Pascal p-value | 0.267 |
| Sherlock p-value | 0.58 |
| Fetal beta | 1.523 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 7 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1976 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18156963 | 1 | 23153308 | EPHB2 | 1.95E-5 | -0.297 | 0.016 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
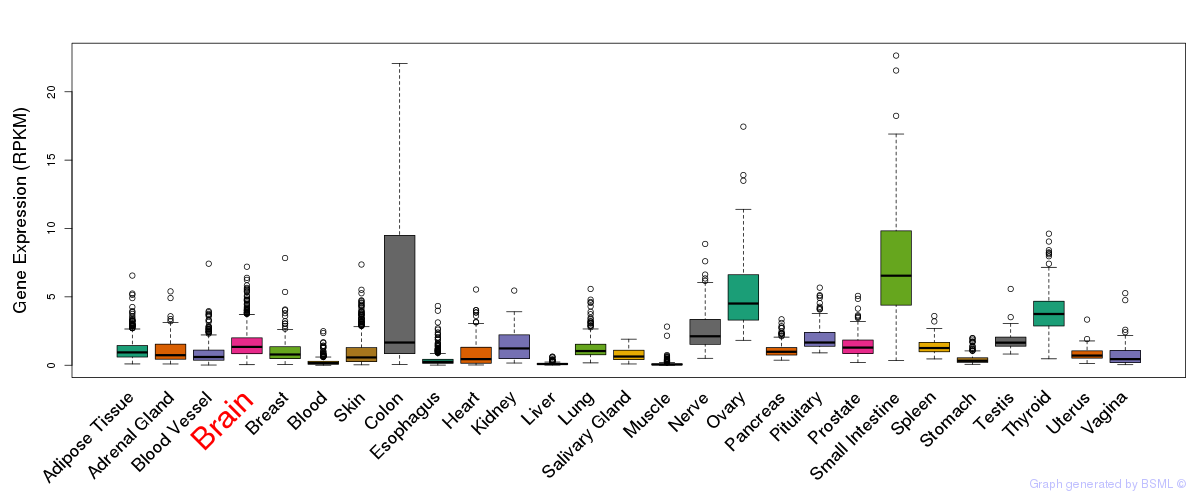
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF444 | 0.83 | 0.87 |
| PRAF2 | 0.82 | 0.83 |
| EPN1 | 0.81 | 0.84 |
| FBXW5 | 0.81 | 0.84 |
| RNF126 | 0.81 | 0.83 |
| ZNF580 | 0.80 | 0.84 |
| PIM3 | 0.80 | 0.85 |
| AC129492.2 | 0.80 | 0.82 |
| C11orf68 | 0.79 | 0.82 |
| DAPK3 | 0.79 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.53 | -0.43 |
| AF347015.8 | -0.49 | -0.41 |
| AF347015.31 | -0.49 | -0.41 |
| MT-CO2 | -0.48 | -0.39 |
| MT-ATP8 | -0.47 | -0.45 |
| AF347015.27 | -0.46 | -0.40 |
| MT-CYB | -0.46 | -0.38 |
| AF347015.33 | -0.45 | -0.38 |
| NOSTRIN | -0.45 | -0.40 |
| AF347015.18 | -0.44 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008046 | axon guidance receptor activity | IEA | axon (GO term level: 6) | - |
| GO:0005102 | receptor binding | IEA | Neurotransmitter (GO term level: 4) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005005 | transmembrane-ephrin receptor activity | TAS | 8589679 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | IEA | neuron, neurogenesis, Synap (GO term level: 11) | - |
| GO:0048168 | regulation of neuronal synaptic plasticity | IEA | neuron, Synap (GO term level: 9) | - |
| GO:0050770 | regulation of axonogenesis | IEA | neuron, axon (GO term level: 13) | - |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 8589679 |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | TAS | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0007612 | learning | IEA | - | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | TAS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 11494128 |
| ACP1 | HAAP | MGC111030 | MGC3499 | acid phosphatase 1, soluble | - | HPRD,BioGRID | 9499402 |
| ADAM17 | CD156b | MGC71942 | TACE | cSVP | ADAM metallopeptidase domain 17 | - | HPRD | 12058067 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Biochemical Activity | BioGRID | 15225643 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Reconstituted Complex | BioGRID | 10644995 |
| EFNB1 | CFND | CFNS | EFL3 | EPLG2 | Elk-L | LERK2 | MGC8782 | ephrin-B1 | - | HPRD | 8878483 |
| EFNB1 | CFND | CFNS | EFL3 | EPLG2 | Elk-L | LERK2 | MGC8782 | ephrin-B1 | The membrane bound ligand ELK-L interacts with the receptor NUK. This interaction was modeled on a demonstrated interaction between ELK-L from human and NUK from mouse. | BIND | 8755474 |
| EFNB2 | EPLG5 | HTKL | Htk-L | LERK5 | MGC126226 | MGC126227 | MGC126228 | ephrin-B2 | The membrane bound ligand HTK-L interacts with the receptor NUK. This interaction was modeled on a demonstrated interaction between HTK-L from human and NUK from mouse. | BIND | 8755474 |
| EFNB2 | EPLG5 | HTKL | Htk-L | LERK5 | MGC126226 | MGC126227 | MGC126228 | ephrin-B2 | - | HPRD,BioGRID | 11780069 |
| EPHB2 | CAPB | DRT | EPHT3 | ERK | Hek5 | MGC87492 | PCBC | Tyro5 | EPH receptor B2 | - | HPRD,BioGRID | 9933164 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 11754835 |
| ITSN1 | ITSN | MGC134948 | MGC134949 | SH3D1A | SH3P17 | intersectin 1 (SH3 domain protein) | - | HPRD,BioGRID | 12389031 |
| MLLT4 | AF-6 | AF6 | AFADIN | FLJ34371 | RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | EphB2 interacts with AF6. This interaction was modelled on a demonstrated interaction between EphB2 from human and mouse AF6. | BIND | 9707552 |
| MLLT4 | AF-6 | AF6 | AFADIN | FLJ34371 | RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | - | HPRD,BioGRID | 9707552 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD | 9233798 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 9233798 |10644995 |
| RP6-213H19.1 | MASK | MST4 | serine/threonine protein kinase MST4 | Phenotypic Enhancement | BioGRID | 11641781 |
| RYK | D3S3195 | JTK5 | JTK5A | RYK1 | RYK receptor-like tyrosine kinase | - | HPRD,BioGRID | 11956217 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 9632142 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| ST GA12 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN | 45 | 34 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS INDUCED BY AKT1 48HR UP | 14 | 8 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 SIGNALING VIA CTNNB1 | 83 | 58 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C4 | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| MCMURRAY TP53 HRAS COOPERATION RESPONSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PLASARI NFIC TARGETS BASAL DN | 18 | 13 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR UP | 33 | 25 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF PERSISTENTLY DN | 61 | 36 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 1497 | 1504 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-150 | 309 | 316 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-185 | 353 | 360 | 1A,m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-203.1 | 1012 | 1018 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| hsa-miR-203 | UGAAAUGUUUAGGACCACUAG | ||||
| miR-224 | 1071 | 1077 | 1A | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-24* | 1076 | 1082 | 1A | hsa-miR-189 | GUGCCUACUGAGCUGAUAUCAGU |
| miR-27 | 1498 | 1504 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 888 | 895 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-448 | 1103 | 1109 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-495 | 885 | 891 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-9 | 1177 | 1183 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.