Gene Page: GATA2
Summary ?
| GeneID | 2624 |
| Symbol | GATA2 |
| Synonyms | DCML|IMD21|MONOMAC|NFE1B |
| Description | GATA binding protein 2 |
| Reference | MIM:137295|HGNC:HGNC:4171|Ensembl:ENSG00000179348|HPRD:00673|Vega:OTTHUMG00000159689 |
| Gene type | protein-coding |
| Map location | 3q21.3 |
| Pascal p-value | 0.098 |
| Fetal beta | -0.758 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.243 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02836487 | 3 | 128206457 | GATA2 | 8.44E-8 | -0.01 | 1.96E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
General gene expression (GTEx)
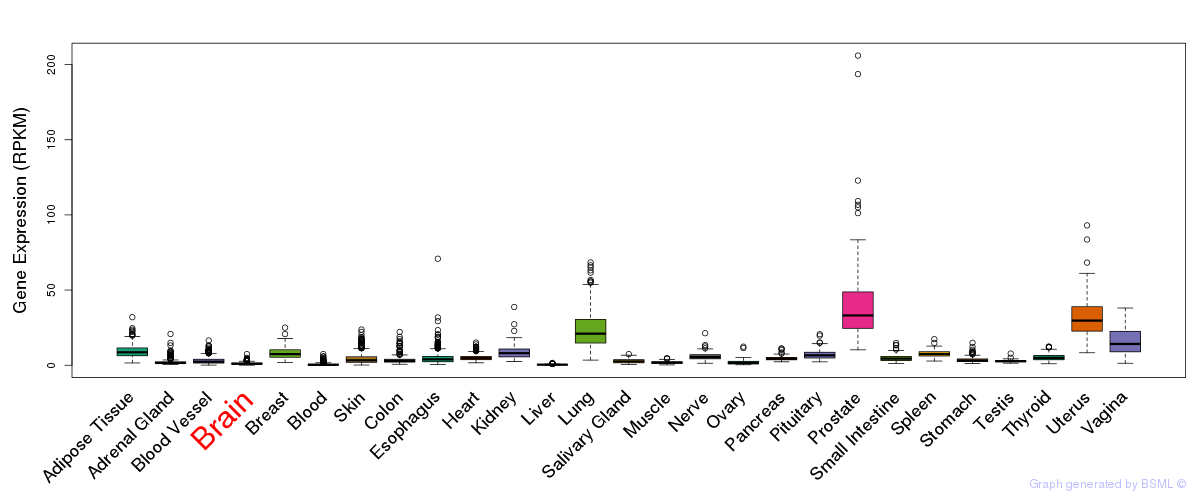
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 8078582 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008134 | transcription factor binding | IPI | 15016828 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030182 | neuron differentiation | IEA | neuron (GO term level: 8) | - |
| GO:0001709 | cell fate determination | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 1370462 | |
| GO:0010552 | positive regulation of specific transcription from RNA polymerase II promoter | IDA | 15016828 | |
| GO:0048469 | cell maturation | IEA | - | |
| GO:0021983 | pituitary gland development | IEA | - | |
| GO:0050766 | positive regulation of phagocytosis | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | TAS | 1370462 | |
| GO:0005730 | nucleolus | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD,BioGRID | 11567998 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | - | HPRD,BioGRID | 11567998 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 11278891 |
| LMO2 | RBTN2 | RBTNL1 | RHOM2 | TTG2 | LIM domain only 2 (rhombotin-like 1) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7568177 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 10938104 |
| POU1F1 | GHF-1 | PIT1 | Pit-1 | POU class 1 homeobox 1 | - | HPRD,BioGRID | 10367888 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | - | HPRD | 10938104 |
| SPI1 | OF | PU.1 | SFPI1 | SPI-1 | SPI-A | spleen focus forming virus (SFFV) proviral integration oncogene spi1 | - | HPRD,BioGRID | 10411939 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | Two-hybrid | BioGRID | 7568177 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 11964310 |
| ZBTB32 | FAXF | FAZF | Rog | TZFP | ZNF538 | zinc finger and BTB domain containing 32 | - | HPRD,BioGRID | 11964310 |
| ZFPM1 | FOG | FOG1 | ZNF408 | ZNF89A | zinc finger protein, multitype 1 | - | HPRD,BioGRID | 12483298 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| SCHRAMM INHBA TARGETS DN | 24 | 12 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 UP | 51 | 30 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| ROSS AML OF FAB M7 TYPE | 68 | 44 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA UP | 22 | 12 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS DN | 30 | 21 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA MAST CELL | 46 | 34 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION DN | 30 | 23 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D4 | 55 | 37 | All SZGR 2.0 genes in this pathway |
| DELLA RESPONSE TO TSA AND BUTYRATE | 21 | 17 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| TIAN TNF SIGNALING NOT VIA NFKB | 22 | 16 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 8 | 49 | 36 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 155 | 161 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-132/212 | 1561 | 1567 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-200bc/429 | 1425 | 1431 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-25/32/92/363/367 | 1336 | 1343 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 155 | 162 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-493-5p | 1496 | 1503 | 1A,m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.