Gene Page: GRIK1
Summary ?
| GeneID | 2897 |
| Symbol | GRIK1 |
| Synonyms | EAA3|EEA3|GLR5|GLUR5|GluK1|gluR-5 |
| Description | glutamate ionotropic receptor kainate type subunit 1 |
| Reference | MIM:138245|HGNC:HGNC:4579|Ensembl:ENSG00000171189|HPRD:00693|Vega:OTTHUMG00000078879 |
| Gene type | protein-coding |
| Map location | 21q22.11 |
| Pascal p-value | 0.132 |
| Sherlock p-value | 0.562 |
| Fetal beta | -3.008 |
| eGene | Myers' cis & trans |
| Support | LIGAND GATED ION SIGNALING Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 9 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0018 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2833869 | chr21 | 33911706 | GRIK1 | 2897 | 0.17 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
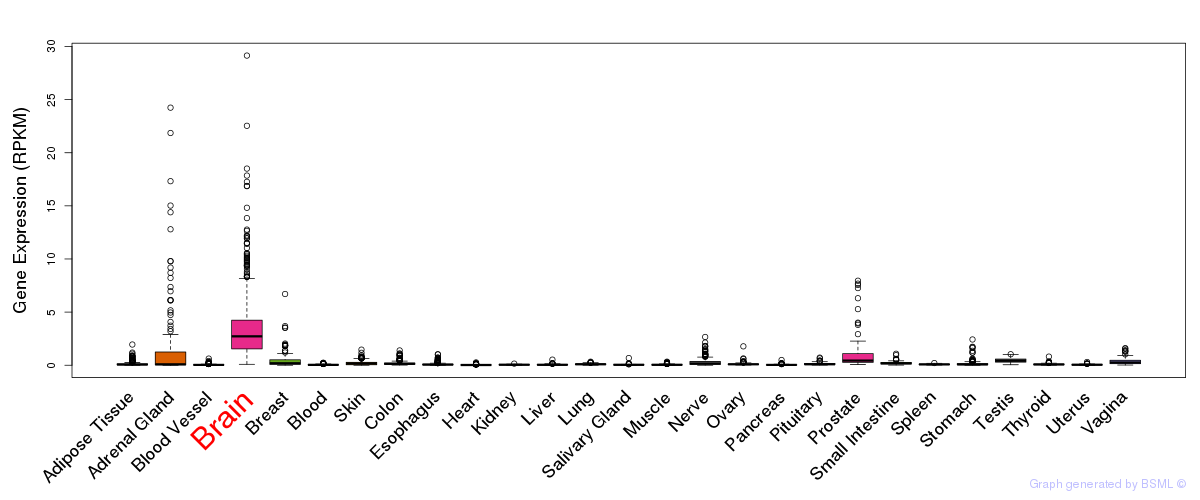
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| INPP5A | 0.81 | 0.80 |
| ACHE | 0.81 | 0.74 |
| B3GALTL | 0.78 | 0.59 |
| RGS8 | 0.77 | 0.74 |
| C1orf127 | 0.76 | 0.51 |
| CACNG5 | 0.75 | 0.58 |
| TTC39A | 0.74 | 0.63 |
| RSPO4 | 0.74 | 0.43 |
| PRKCH | 0.74 | 0.43 |
| PCBP3 | 0.74 | 0.65 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.27 | -0.35 |
| AF347015.2 | -0.26 | -0.29 |
| EMX2 | -0.25 | -0.38 |
| PLAC9 | -0.24 | -0.30 |
| AL139819.3 | -0.24 | -0.39 |
| RPL31 | -0.24 | -0.26 |
| GNG11 | -0.24 | -0.39 |
| AF347015.31 | -0.24 | -0.31 |
| RLBP1 | -0.23 | -0.27 |
| FAM159B | -0.23 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005234 | extracellular-glutamate-gated ion channel activity | IEA | glutamate (GO term level: 11) | - |
| GO:0015277 | kainate selective glutamate receptor activity | IDA | glutamate (GO term level: 8) | 9335499 |
| GO:0015277 | kainate selective glutamate receptor activity | IEA | glutamate (GO term level: 8) | - |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005261 | cation channel activity | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 10580501 |
| GO:0051966 | regulation of synaptic transmission, glutamatergic | TAS | neuron, glutamate, Synap, Neurotransmitter (GO term level: 9) | 9335499 |
| GO:0007268 | synaptic transmission | IEA | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 10580501 |
| GO:0007215 | glutamate signaling pathway | TAS | glutamate (GO term level: 7) | 8260617 |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8260617 | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| DORSEY GAB2 TARGETS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |