Gene Page: IL10
Summary ?
| GeneID | 3586 |
| Symbol | IL10 |
| Synonyms | CSIF|GVHDS|IL-10|IL10A|TGIF |
| Description | interleukin 10 |
| Reference | MIM:124092|HGNC:HGNC:5962|Ensembl:ENSG00000136634|HPRD:00495|Vega:OTTHUMG00000036386 |
| Gene type | protein-coding |
| Map location | 1q31-q32 |
| Fetal beta | -0.241 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 3 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
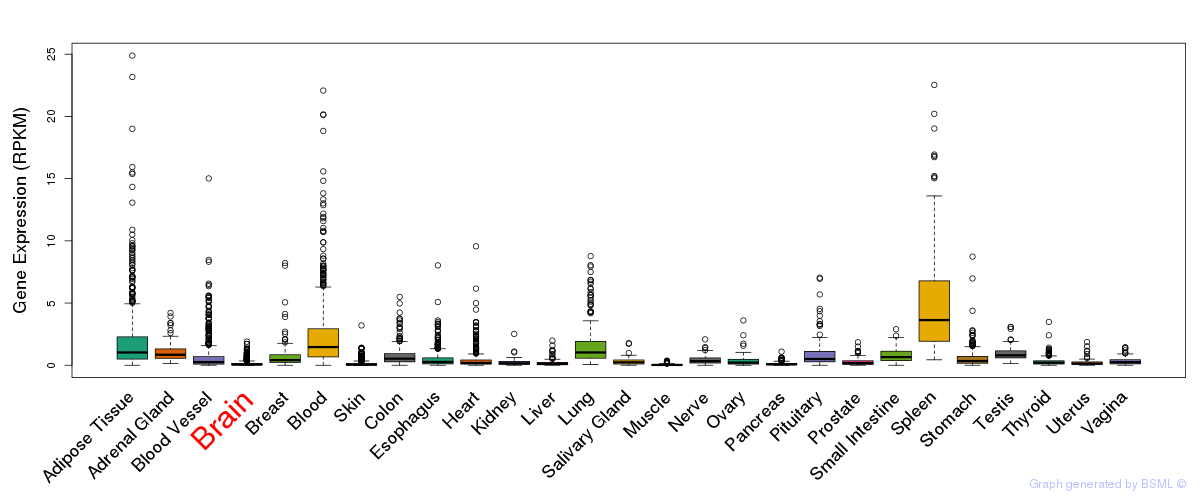
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRPL11 | 0.81 | 0.73 |
| MPDU1 | 0.79 | 0.78 |
| HADH | 0.79 | 0.73 |
| SNAPIN | 0.78 | 0.75 |
| C17orf81 | 0.78 | 0.78 |
| STOML2 | 0.76 | 0.70 |
| ADSL | 0.76 | 0.70 |
| VKORC1 | 0.75 | 0.78 |
| MTX1 | 0.75 | 0.74 |
| GABARAP | 0.75 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.46 | -0.49 |
| AF347015.18 | -0.45 | -0.54 |
| AF347015.8 | -0.45 | -0.48 |
| MT-ATP8 | -0.45 | -0.53 |
| AF347015.26 | -0.45 | -0.50 |
| MT-CYB | -0.44 | -0.47 |
| AF347015.15 | -0.42 | -0.47 |
| MT-CO2 | -0.42 | -0.44 |
| AF347015.33 | -0.41 | -0.45 |
| AF347015.2 | -0.41 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005141 | interleukin-10 receptor binding | NAS | 1847510 | |
| GO:0005125 | cytokine activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 11485736 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0032800 | receptor biosynthetic process | IDA | Neurotransmitter (GO term level: 5) | 10443688 |
| GO:0030886 | negative regulation of myeloid dendritic cell activation | IEA | dendrite (GO term level: 8) | - |
| GO:0002740 | negative regulation of cytokine secretion during immune response | IDA | 10443688 | |
| GO:0002237 | response to molecule of bacterial origin | IDA | 17449476 | |
| GO:0007267 | cell-cell signaling | IC | 1847510 | |
| GO:0007253 | cytoplasmic sequestering of NF-kappaB | NAS | 10975994 | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006954 | inflammatory response | NAS | 9463379 | |
| GO:0006916 | anti-apoptosis | NAS | 8312229 | |
| GO:0010468 | regulation of gene expression | IDA | 9184696 | |
| GO:0042100 | B cell proliferation | NAS | 8228801 | |
| GO:0030595 | leukocyte chemotaxis | TAS | 9405662 | |
| GO:0042130 | negative regulation of T cell proliferation | NAS | 8499633 | |
| GO:0042742 | defense response to bacterium | IEA | - | |
| GO:0032695 | negative regulation of interleukin-12 production | IEA | - | |
| GO:0042092 | T-helper 2 type immune response | TAS | 11244051 | |
| GO:0032715 | negative regulation of interleukin-6 production | IDA | 10443688 | |
| GO:0030097 | hemopoiesis | TAS | 11244051 | |
| GO:0030183 | B cell differentiation | NAS | 8228801 | |
| GO:0045191 | regulation of isotype switching | NAS | 8228801 | |
| GO:0050715 | positive regulation of cytokine secretion | IDA | 10443688 | |
| GO:0045347 | negative regulation of MHC class II biosynthetic process | TAS | 11244051 | |
| GO:0045348 | positive regulation of MHC class II biosynthetic process | IEA | - | |
| GO:0045355 | negative regulation of interferon-alpha biosynthetic process | NAS | 9637497 | |
| GO:0051384 | response to glucocorticoid stimulus | IDA | 10443688 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | NAS | 1847510 | |
| GO:0005615 | extracellular space | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG INTESTINAL IMMUNE NETWORK FOR IGA PRODUCTION | 48 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA | 30 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG AUTOIMMUNE THYROID DISEASE | 53 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ASBCELL PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DC PATHWAY | 22 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL10 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 1 | 13 | 11 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 1Q32 AMPLICON | 13 | 5 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA UP | 51 | 29 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS | 54 | 30 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| GAVIN PDE3B TARGETS | 22 | 15 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| BUDHU LIVER CANCER METASTASIS UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SEIKE LUNG CANCER POOR SURVIVAL | 11 | 8 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS AND CYCLIC RGD | 21 | 15 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY DN | 14 | 11 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 140 | 146 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-27 | 178 | 184 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-361 | 135 | 141 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-369-3p | 205 | 211 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 205 | 212 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.