Gene Page: SMAD7
Summary ?
| GeneID | 4092 |
| Symbol | SMAD7 |
| Synonyms | CRCS3|MADH7|MADH8 |
| Description | SMAD family member 7 |
| Reference | MIM:602932|HGNC:HGNC:6773|Ensembl:ENSG00000101665|HPRD:04241|Vega:OTTHUMG00000132655 |
| Gene type | protein-coding |
| Map location | 18q21.1 |
| Pascal p-value | 0.816 |
| Sherlock p-value | 0.247 |
| TADA p-value | 0.009 |
| Fetal beta | -0.589 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SMAD7 | chr18 | 46474778 | G | C | NM_001190821 NM_001190822 NM_005904 | p.215P>A . p.215P>A | missense 5-UTR missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4405616 | chr18 | 45593097 | SMAD7 | 4092 | 0.14 | cis |
Section II. Transcriptome annotation
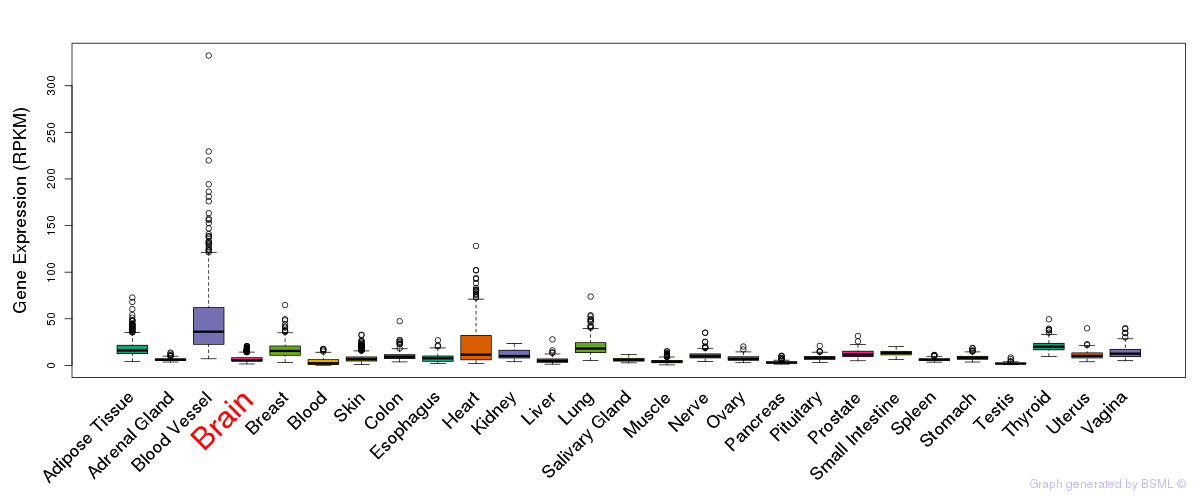
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR78 | 0.89 | 0.38 |
| LRP2BP | 0.88 | 0.44 |
| YSK4 | 0.88 | 0.21 |
| CCDC81 | 0.88 | 0.35 |
| SPAG8 | 0.88 | 0.60 |
| WDR63 | 0.88 | 0.18 |
| CCDC108 | 0.88 | 0.37 |
| C7orf57 | 0.88 | 0.30 |
| ARMC3 | 0.88 | 0.33 |
| RSHL1 | 0.87 | 0.36 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.22 | -0.29 |
| AF347015.15 | -0.21 | -0.27 |
| AF347015.31 | -0.20 | -0.27 |
| MT-CYB | -0.20 | -0.27 |
| AF347015.2 | -0.20 | -0.28 |
| AF347015.8 | -0.20 | -0.25 |
| MT-CO2 | -0.19 | -0.24 |
| MT-ATP8 | -0.19 | -0.28 |
| AF347015.33 | -0.19 | -0.22 |
| GBP2 | -0.18 | -0.24 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVR1B | ACTRIB | ACVRLK4 | ALK4 | SKR2 | activin A receptor, type IB | - | HPRD,BioGRID | 12023024 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Affinity Capture-Western | BioGRID | 16362038 |
| BMPR1A | 10q23del | ACVRLK3 | ALK3 | CD292 | bone morphogenetic protein receptor, type IA | Affinity Capture-Western | BioGRID | 15148321 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western Co-localization | BioGRID | 15684397 |
| DVL1 | DVL | MGC54245 | dishevelled, dsh homolog 1 (Drosophila) | Reconstituted Complex | BioGRID | 12650946 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 12408818 |
| ERBB2IP | ERBIN | LAP2 | erbb2 interacting protein | - | HPRD,BioGRID | 12650946 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | Affinity Capture-Western | BioGRID | 15684397 |
| MAP2K3 | MAPKK3 | MEK3 | MKK3 | PRKMK3 | mitogen-activated protein kinase kinase 3 | - | HPRD,BioGRID | 12589052 |
| MAP2K6 | MAPKK6 | MEK6 | MKK6 | PRKMK6 | SAPKK3 | mitogen-activated protein kinase kinase 6 | - | HPRD,BioGRID | 12589052 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD,BioGRID | 12589052 |
| MAP3K7IP1 | 3'-Tab1 | MGC57664 | TAB1 | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | - | HPRD,BioGRID | 11737269 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | - | HPRD,BioGRID | 12589052 |
| PARD3 | ASIP | Baz | Bazooka | FLJ21015 | PAR3 | PAR3alpha | PARD3A | SE2-5L16 | SE2-5LT1 | SE2-5T2 | par-3 partitioning defective 3 homolog (C. elegans) | - | HPRD | 12650946 |
| PARD3B | ALS2CR19 | MGC16131 | PAR3B | PAR3L | PAR3LC | PAR3beta | Par3Lb | par-3 partitioning defective 3 homolog B (C. elegans) | Reconstituted Complex | BioGRID | 12650946 |
| PIAS4 | FLJ12419 | MGC35296 | PIASY | Piasg | ZMIZ6 | protein inhibitor of activated STAT, 4 | - | HPRD,BioGRID | 12815042 |
| RNF111 | ARK | DKFZp313E0731 | DKFZp686H1966 | DKFZp761D081 | FLJ38008 | ring finger protein 111 | - | HPRD,BioGRID | 14657019 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Phenotypic Suppression | BioGRID | 9892009 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Phenotypic Suppression | BioGRID | 9892009 |10757800 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Phenotypic Suppression | BioGRID | 9892009 |
| SMAD6 | HsT17432 | MADH6 | MADH7 | SMAD family member 6 | - | HPRD,BioGRID | 9256479 |
| SMURF1 | KIAA1625 | SMAD specific E3 ubiquitin protein ligase 1 | - | HPRD | 11278251 |12151385 |12519765 |
| SMURF1 | KIAA1625 | SMAD specific E3 ubiquitin protein ligase 1 | Affinity Capture-Western in vitro in vivo | BioGRID | 11278251 |12151385 |14722617 |
| SMURF1 | KIAA1625 | SMAD specific E3 ubiquitin protein ligase 1 | Smad7 interacts with Smurf1. This interaction was modeled on a demonstrated interaction between Smad7 and Smurf1, both from an unspecified species. | BIND | 15820682 |
| SMURF2 | DKFZp686F0270 | MGC138150 | SMAD specific E3 ubiquitin protein ligase 2 | - | HPRD,BioGRID | 11163210 |
| SMURF2 | DKFZp686F0270 | MGC138150 | SMAD specific E3 ubiquitin protein ligase 2 | SMAD7 interacts with SMURF2. | BIND | 15761153 |
| STAMBP | AMSH | MGC126516 | MGC126518 | STAM binding protein | - | HPRD,BioGRID | 11483516 |
| STRAP | MAWD | PT-WD | UNRIP | serine/threonine kinase receptor associated protein | - | HPRD,BioGRID | 10757800 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD,BioGRID | 9215638 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | - | HPRD,BioGRID | 11163210 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | - | HPRD,BioGRID | 12118366 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TGFB PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY BMP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | 23 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY DN | 74 | 44 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| MARKS ACETYLATED NON HISTONE PROTEINS | 15 | 9 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8 | 36 | 28 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA EOSINOPHIL | 30 | 20 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| DORSEY GAB2 TARGETS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| GAUTSCHI SRC SIGNALING | 8 | 6 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE UP | 109 | 68 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 1HR UP | 34 | 26 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |