Gene Page: ASCL1
Summary ?
| GeneID | 429 |
| Symbol | ASCL1 |
| Synonyms | ASH1|HASH1|MASH1|bHLHa46 |
| Description | achaete-scute family bHLH transcription factor 1 |
| Reference | MIM:100790|HGNC:HGNC:738|Ensembl:ENSG00000139352|HPRD:00011|Vega:OTTHUMG00000169967 |
| Gene type | protein-coding |
| Map location | 12q23.2 |
| Pascal p-value | 0.004 |
| Fetal beta | 0.91 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 7 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
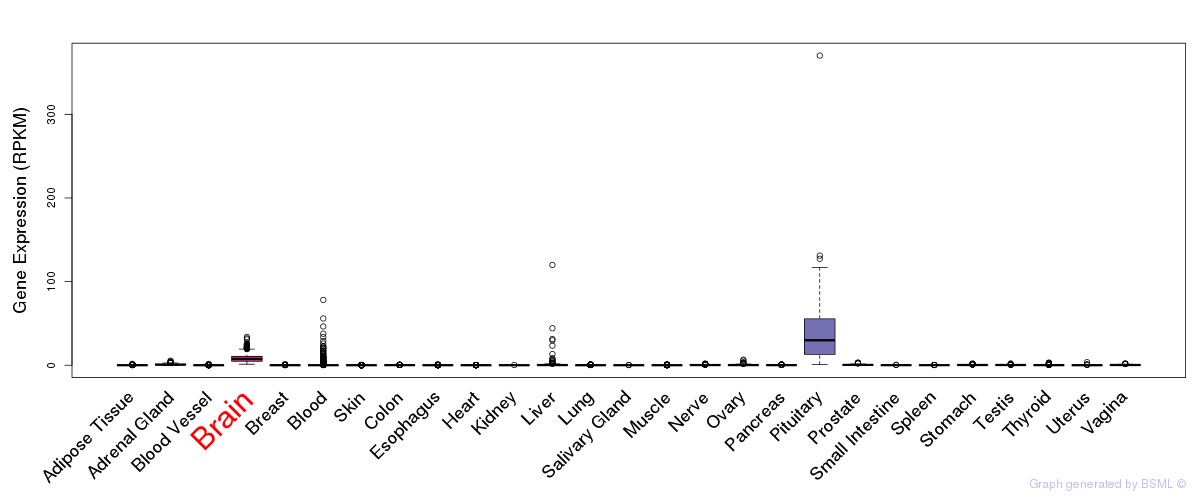
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NT5DC2 | 0.90 | 0.83 |
| SIRT6 | 0.90 | 0.90 |
| MIIP | 0.90 | 0.87 |
| ACTL6B | 0.90 | 0.90 |
| SHF | 0.90 | 0.86 |
| DCAF15 | 0.90 | 0.86 |
| FAM110A | 0.89 | 0.86 |
| ZNF821 | 0.89 | 0.90 |
| AGBL5 | 0.89 | 0.89 |
| DOK4 | 0.89 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.72 | -0.76 |
| AF347015.27 | -0.71 | -0.85 |
| AF347015.31 | -0.69 | -0.83 |
| MT-CO2 | -0.69 | -0.84 |
| AF347015.33 | -0.69 | -0.83 |
| MT-CYB | -0.68 | -0.84 |
| AIFM3 | -0.67 | -0.68 |
| S100B | -0.67 | -0.75 |
| C5orf53 | -0.67 | -0.65 |
| AF347015.8 | -0.67 | -0.85 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | NAS | 8390674 | |
| GO:0005515 | protein binding | IPI | 10903890 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060165 | regulation of timing of subpallium neuron differentiation | IEA | neuron (GO term level: 11) | - |
| GO:0060163 | subpallium neuron fate commitment | IEA | neuron (GO term level: 11) | - |
| GO:0007405 | neuroblast proliferation | IEA | neuron (GO term level: 8) | - |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0048666 | neuron development | IEA | neuron (GO term level: 9) | - |
| GO:0010001 | glial cell differentiation | IEA | Glial (GO term level: 8) | - |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007219 | Notch signaling pathway | IEA | - | |
| GO:0007400 | neuroblast fate determination | IEA | - | |
| GO:0007389 | pattern specification process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| FRASOR TAMOXIFEN RESPONSE UP | 51 | 36 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP | 71 | 48 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS UP | 43 | 28 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-22 | 1020 | 1026 | 1A | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-369-3p | 669 | 675 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 669 | 676 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-375 | 1007 | 1013 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-493-5p | 771 | 777 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-496 | 811 | 817 | m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.