Gene Page: NUDT1
Summary ?
| GeneID | 4521 |
| Symbol | NUDT1 |
| Synonyms | MTH1 |
| Description | nudix hydrolase 1 |
| Reference | MIM:600312|HGNC:HGNC:8048|Ensembl:ENSG00000106268|HPRD:02634|Vega:OTTHUMG00000023072 |
| Gene type | protein-coding |
| Map location | 7p22 |
| Pascal p-value | 6.171E-5 |
| Sherlock p-value | 0.596 |
| Fetal beta | 1.401 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6792339 | chr3 | 99680906 | NUDT1 | 4521 | 0.17 | trans | ||
| rs7830259 | chr8 | 14087643 | NUDT1 | 4521 | 0.07 | trans |
Section II. Transcriptome annotation
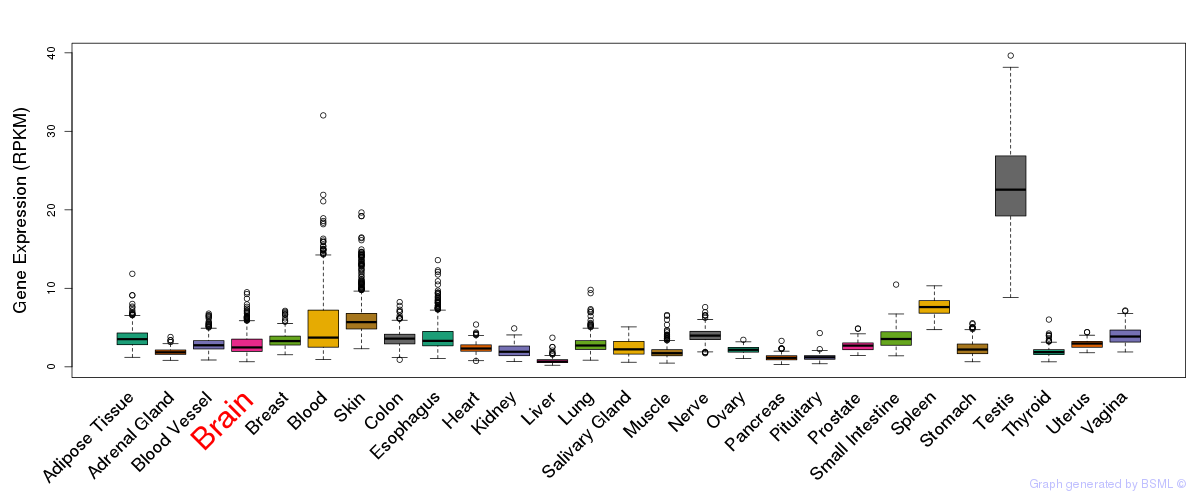
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IQGAP2 | 0.80 | 0.44 |
| TFPI | 0.78 | 0.21 |
| EEPD1 | 0.78 | 0.59 |
| CNGA3 | 0.78 | 0.43 |
| ANKFN1 | 0.76 | 0.52 |
| TGIF1 | 0.75 | 0.41 |
| TNC | 0.75 | 0.39 |
| RASSF10 | 0.74 | 0.29 |
| CDO1 | 0.74 | 0.52 |
| VCAM1 | 0.74 | 0.43 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.27 | -0.29 |
| MT-CO2 | -0.27 | -0.28 |
| AF347015.8 | -0.26 | -0.27 |
| MT-CYB | -0.26 | -0.27 |
| COX4I2 | -0.25 | -0.28 |
| AF347015.27 | -0.25 | -0.25 |
| AF347015.33 | -0.25 | -0.25 |
| CXCL14 | -0.25 | -0.27 |
| AF347015.21 | -0.24 | -0.31 |
| AF347015.2 | -0.24 | -0.25 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7P22 AMPLICON | 38 | 27 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |