Gene Page: ATM
Summary ?
| GeneID | 472 |
| Symbol | ATM |
| Synonyms | AT1|ATA|ATC|ATD|ATDC|ATE|TEL1|TELO1 |
| Description | ATM serine/threonine kinase |
| Reference | MIM:607585|HGNC:HGNC:795|Ensembl:ENSG00000149311|HPRD:06347|Vega:OTTHUMG00000166480 |
| Gene type | protein-coding |
| Map location | 11q22-q23 |
| Pascal p-value | 0.768 |
| Sherlock p-value | 0.543 |
| Fetal beta | 1.15 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0817 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
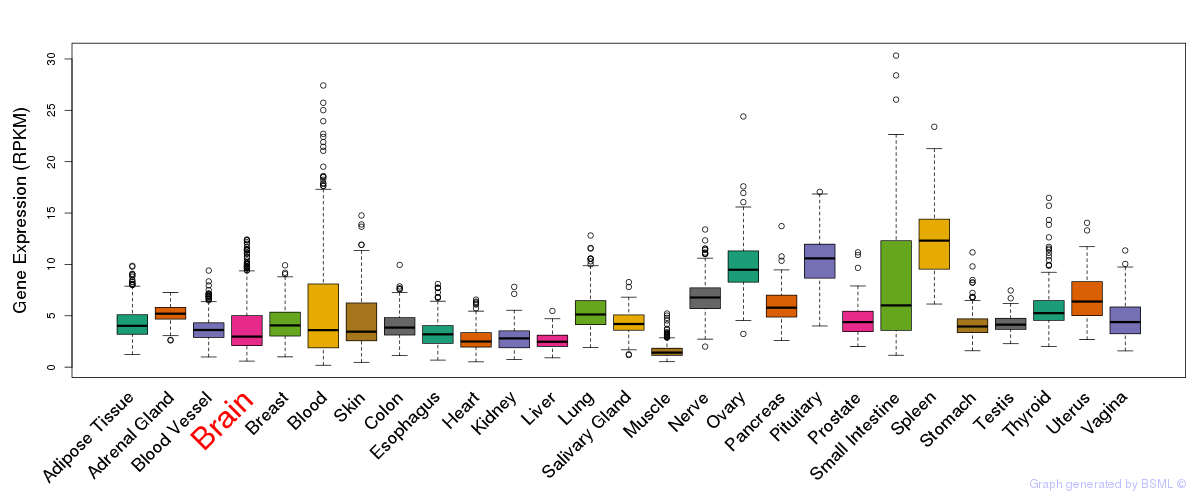
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ROBLD3 | 0.88 | 0.85 |
| CCDC56 | 0.85 | 0.81 |
| MRPL36 | 0.85 | 0.78 |
| PRELID1 | 0.83 | 0.77 |
| PSMB3 | 0.82 | 0.80 |
| RNF181 | 0.82 | 0.83 |
| MRPL20 | 0.81 | 0.73 |
| GABARAP | 0.81 | 0.77 |
| PSMB6 | 0.81 | 0.84 |
| RNF7 | 0.81 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-ATP8 | -0.42 | -0.46 |
| AF347015.18 | -0.40 | -0.44 |
| COBLL1 | -0.40 | -0.40 |
| PTRF | -0.40 | -0.42 |
| MYH9 | -0.39 | -0.37 |
| AF347015.26 | -0.39 | -0.43 |
| EPAS1 | -0.39 | -0.41 |
| GIMAP8 | -0.38 | -0.40 |
| AF347015.2 | -0.38 | -0.40 |
| FLT1 | -0.37 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IDA | 11375976 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016303 | 1-phosphatidylinositol-3-kinase activity | IMP | 11375976 | |
| GO:0047485 | protein N-terminus binding | IDA | 11375976 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0051402 | neuron apoptosis | IEA | neuron, axon, dendrite (GO term level: 7) | - |
| GO:0000077 | DNA damage checkpoint | IEA | - | |
| GO:0001756 | somitogenesis | IEA | - | |
| GO:0007131 | reciprocal meiotic recombination | TAS | 7792600 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0008219 | cell death | IEA | - | |
| GO:0007292 | female gamete generation | IEA | - | |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0006281 | DNA repair | TAS | 9733515 | |
| GO:0007165 | signal transduction | TAS | 7792600 | |
| GO:0008630 | DNA damage response, signal transduction resulting in induction of apoptosis | IEA | - | |
| GO:0007094 | mitotic cell cycle spindle assembly checkpoint | IMP | 11943150 | |
| GO:0007507 | heart development | IEA | - | |
| GO:0010212 | response to ionizing radiation | IDA | 11375976 | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005819 | spindle | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 10550055 |10959836 |11331603 |12556884 |12607003 | |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 9168117 |
| AP1B1 | ADTB1 | AP105A | BAM22 | CLAPB2 | adaptor-related protein complex 1, beta 1 subunit | Protein-peptide | BioGRID | 10608806 |
| AP2A2 | ADTAB | CLAPA2 | HIP9 | HYPJ | adaptor-related protein complex 2, alpha 2 subunit | Protein-peptide | BioGRID | 10608806 |
| AP2B1 | ADTB2 | AP105B | AP2-BETA | CLAPB1 | DKFZp781K0743 | adaptor-related protein complex 2, beta 1 subunit | - | HPRD | 9707615 |
| AP3B2 | DKFZp686D17136 | NAPTB | adaptor-related protein complex 3, beta 2 subunit | - | HPRD | 9707615 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | An unspecified isoform of ATM interacts with and phosphorylates ATF2. | BIND | 15916964 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Protein-peptide | BioGRID | 10608806 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | ATM interacts with itself to form a dimer. | BIND | 15790808 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Protein-peptide | BioGRID | 10608806 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 10783165 |12034743 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-Western Co-purification Protein-peptide Reconstituted Complex | BioGRID | 10550055 |10608806 |10783165 |10866324 |11016625 |11114888 |11278964 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 2001833 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | Reconstituted Complex | BioGRID | 15199141 |
| CAT | MGC138422 | MGC138424 | catalase | - | HPRD | 10567403 |
| CHEK1 | CHK1 | CHK1 checkpoint homolog (S. pombe) | Protein-peptide | BioGRID | 10608806 |
| CHEK2 | CDS1 | CHK2 | HuCds1 | LFS2 | PP1425 | RAD53 | CHK2 checkpoint homolog (S. pombe) | ATM phosphorylates Chk2 at threonine 68. | BIND | 15790808 |
| CHEK2 | CDS1 | CHK2 | HuCds1 | LFS2 | PP1425 | RAD53 | CHK2 checkpoint homolog (S. pombe) | ATM phosphorylates Chk2 at threonine 68. | BIND | 15064416 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | Affinity Capture-Western | BioGRID | 15073328 |
| E4F1 | E4F | MGC99614 | E4F transcription factor 1 | Protein-peptide | BioGRID | 10608806 |
| EEF1E1 | AIMP3 | P18 | eukaryotic translation elongation factor 1 epsilon 1 | p18 interacts with ATM. | BIND | 15680327 |
| EIF4EBP1 | 4E-BP1 | 4EBP1 | BP-1 | MGC4316 | PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 | Protein-peptide | BioGRID | 10608806 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12086603 |14499622 |
| H2AFX | H2A.X | H2A/X | H2AX | H2A histone family, member X | ATM phosphorylates the carboxy terminus of H2AX. | BIND | 15064416 |
| HINT1 | FLJ30414 | FLJ32340 | HINT | PKCI-1 | PRKCNH1 | histidine triad nucleotide binding protein 1 | - | HPRD | 7644499 |
| LIG4 | - | ligase IV, DNA, ATP-dependent | Protein-peptide | BioGRID | 10608806 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | Biochemical Activity | BioGRID | 15210935 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | - | HPRD | 12607005 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | Affinity Capture-Western | BioGRID | 14519663 |
| MLH1 | COCA2 | FCC2 | HNPCC | HNPCC2 | MGC5172 | hMLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | Co-purification | BioGRID | 10783165 |
| MRE11A | ATLD | HNGS1 | MRE11 | MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | ATM interacts with MRE11A (Mre11). | BIND | 15758953 |
| MRE11A | ATLD | HNGS1 | MRE11 | MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | Co-purification Protein-peptide | BioGRID | 10608806 |10783165 |
| MSH2 | COCA1 | FCC1 | HNPCC | HNPCC1 | LCFS2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | Co-purification | BioGRID | 10783165 |
| MSH6 | GTBP | HNPCC5 | HSAP | mutS homolog 6 (E. coli) | Co-purification | BioGRID | 10783165 |
| NBN | AT-V1 | AT-V2 | ATV | FLJ10155 | MGC87362 | NBS | NBS1 | P95 | nibrin | Co-purification Protein-peptide | BioGRID | 10608806 |10783165 |
| NBN | AT-V1 | AT-V2 | ATV | FLJ10155 | MGC87362 | NBS | NBS1 | P95 | nibrin | NBS1 interacts with ATM. | BIND | 15758953 |
| PEX5 | FLJ50634 | FLJ50721 | FLJ51948 | PTS1-BP | PTS1R | PXR1 | peroxisomal biogenesis factor 5 | - | HPRD,BioGRID | 10567403 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD,BioGRID | 10464290 |
| PTS | FLJ97081 | PTPS | 6-pyruvoyltetrahydropterin synthase | Protein-peptide | BioGRID | 10608806 |
| RAD17 | CCYC | FLJ41520 | HRAD17 | R24L | RAD17SP | RAD24 | RAD17 homolog (S. pombe) | - | HPRD,BioGRID | 11418864 |
| RAD50 | RAD50-2 | hRad50 | RAD50 homolog (S. cerevisiae) | Co-purification | BioGRID | 10783165 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | Affinity Capture-Western | BioGRID | 10212258 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | Affinity Capture-Western Protein-peptide Reconstituted Complex | BioGRID | 10608806 |10910365 |
| RFC1 | A1 | MGC51786 | MHCBFB | PO-GA | RECC1 | RFC | RFC140 | replication factor C (activator 1) 1, 145kDa | Co-purification | BioGRID | 10783165 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | Affinity Capture-Western Reconstituted Complex | BioGRID | 15854902 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | ATM interacts with Rheb. | BIND | 15854902 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | Affinity Capture-Western Reconstituted Complex | BioGRID | 11877376 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | - | HPRD,BioGRID | 11375976 |
| TIPARP | DDF1 | DKFZp434J214 | DKFZp686N0351 | DKFZp686P1838 | FLJ40466 | PARP-1 | PARP-7 | PARP7 | TCDD-inducible poly(ADP-ribose) polymerase | - | HPRD | 11238919 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9843217 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | ATM phosphorylates p53 at serine 15. | BIND | 15064416 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | An unspecified isoform of ATM interacts with and phosphorylates p53. This interaction was modeled on a demonstrated interaction between human ATM and p53 from an unspecified species. | BIND | 15916964 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | ATM phosphorylates p53 at Ser15. | BIND | 15175154 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | ATM phosphorylates p53 at serine 15. | BIND | 15790808 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Biochemical Activity Reconstituted Complex | BioGRID | 12447390 |12697768 |
| UPF1 | FLJ43809 | FLJ46894 | HUPF1 | KIAA0221 | NORF1 | RENT1 | pNORF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | ATM phosphorylates hUpf1. | BIND | 15175154 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | Protein-peptide | BioGRID | 10608806 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | XRCC5 (Ku80) interacts with ATM. | BIND | 15758953 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATM PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53 PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RB PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATRBRCA PATHWAY | 21 | 12 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY | 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID NFKAPPAB CANONICAL PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PID BARD1 PATHWAY | 29 | 19 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | 51 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | 17 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME FANCONI ANEMIA PATHWAY | 25 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR | 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | 51 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M CHECKPOINTS | 45 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M DNA DAMAGE CHECKPOINT | 12 | 8 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA DN | 70 | 38 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| WIKMAN ASBESTOS LUNG CANCER DN | 28 | 13 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP DN | 35 | 20 | All SZGR 2.0 genes in this pathway |
| RAMJAUN APOPTOSIS BY TGFB1 VIA SMAD4 DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 23 | 24 | 13 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| SCHRAMM INHBA TARGETS DN | 24 | 12 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS RESISTANT UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 11Q23 DELETION | 23 | 18 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| GALI TP53 TARGETS APOPTOTIC UP | 8 | 7 | All SZGR 2.0 genes in this pathway |
| MATZUK MEIOTIC AND DNA REPAIR | 39 | 26 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE DN | 57 | 36 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS DN | 89 | 50 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-203.1 | 2297 | 2303 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.