Gene Page: PIK3R1
Summary ?
| GeneID | 5295 |
| Symbol | PIK3R1 |
| Synonyms | AGM7|GRB1|IMD36|p85|p85-ALPHA |
| Description | phosphoinositide-3-kinase regulatory subunit 1 |
| Reference | MIM:171833|HGNC:HGNC:8979|Ensembl:ENSG00000145675|HPRD:01381|Vega:OTTHUMG00000131251 |
| Gene type | protein-coding |
| Map location | 5q13.1 |
| Pascal p-value | 0.028 |
| Sherlock p-value | 0.578 |
| Fetal beta | -2.444 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 9.9284 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26076905 | 5 | 67522298 | PIK3R1 | 3.44E-4 | 0.006 | 0.191 | DMG:Montano_2016 |
| cg15021292 | 5 | 67521596 | PIK3R1 | 8.69E-5 | 0.444 | 0.026 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
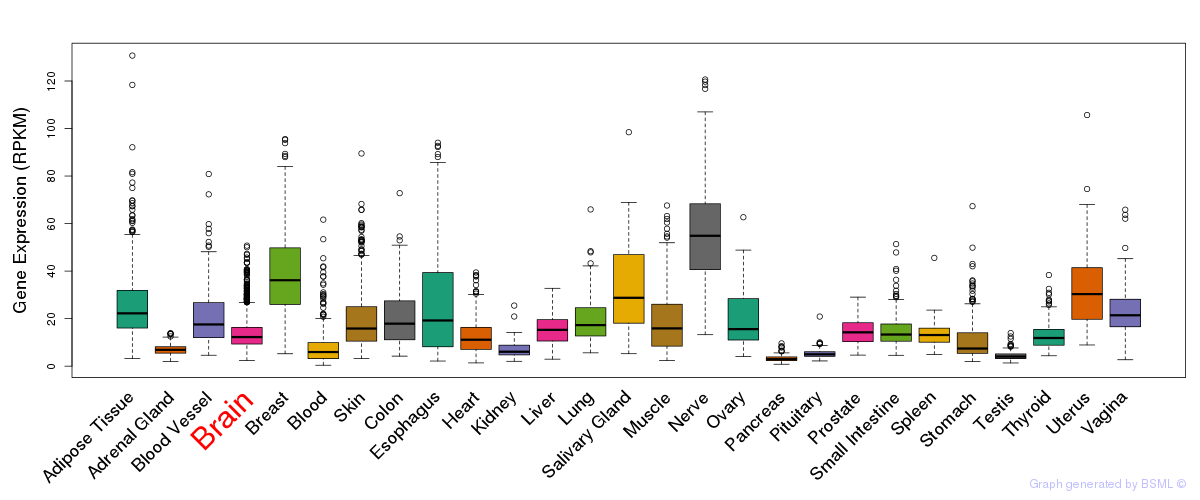
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLCO2B1 | 0.76 | 0.81 |
| LAMB2 | 0.74 | 0.75 |
| MMRN2 | 0.72 | 0.78 |
| TGM2 | 0.71 | 0.79 |
| SNX33 | 0.71 | 0.76 |
| TLN1 | 0.70 | 0.74 |
| ENG | 0.69 | 0.74 |
| TGFBR2 | 0.68 | 0.75 |
| A2M | 0.68 | 0.74 |
| ANGPTL2 | 0.68 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| OXSM | -0.48 | -0.55 |
| NDUFAF2 | -0.46 | -0.54 |
| COMMD3 | -0.45 | -0.54 |
| COQ3 | -0.44 | -0.50 |
| ZNF32 | -0.44 | -0.54 |
| SNHG12 | -0.43 | -0.50 |
| C12orf45 | -0.43 | -0.54 |
| FAM92A1 | -0.43 | -0.47 |
| SLC10A5 | -0.42 | -0.44 |
| GPR22 | -0.42 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005159 | insulin-like growth factor receptor binding | IPI | 7541045 | |
| GO:0005158 | insulin receptor binding | IPI | 7537849 |8276809 | |
| GO:0005545 | phosphatidylinositol binding | NAS | - | |
| GO:0035014 | phosphoinositide 3-kinase regulator activity | ISS | - | |
| GO:0019903 | protein phosphatase binding | IPI | 14699157 | |
| GO:0043125 | ErbB-3 class receptor binding | IDA | 10572067 | |
| GO:0043559 | insulin binding | IDA | 8440175 | |
| GO:0043560 | insulin receptor substrate binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007242 | intracellular signaling cascade | NAS | - | |
| GO:0008286 | insulin receptor signaling pathway | IPI | 8276809 | |
| GO:0048009 | insulin-like growth factor receptor signaling pathway | IPI | 7541045 | |
| GO:0046854 | phosphoinositide phosphorylation | ISS | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9356464 |10648629 |11606067 |12167717 |12660731 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005943 | 1-phosphatidylinositol-4-phosphate 3-kinase, class IA complex | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | c-Abl interacts with p85. This interaction was modeled on a demonstrated interaction between human c-Abl and bovine p85. | BIND | 1383690 |
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Reconstituted Complex | BioGRID | 8294442 |
| ADAM12 | MCMP | MCMPMltna | MLTN | MLTNA | ADAM metallopeptidase domain 12 | - | HPRD,BioGRID | 11313349 |
| AGAP2 | CENTG1 | FLJ16430 | GGAP2 | KIAA0167 | PIKE | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | - | HPRD,BioGRID | 11136977 |
| ARAF | A-RAF | ARAF1 | PKS2 | RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog | - | HPRD,BioGRID | 11812000 |
| ARHGAP1 | CDC42GAP | RHOGAP | RHOGAP1 | p50rhoGAP | Rho GTPase activating protein 1 | - | HPRD,BioGRID | 8253717 |
| ARHGAP17 | DKFZp564A1363 | FLJ37567 | FLJ43368 | MGC87805 | MST066 | MST110 | MSTP038 | MSTP066 | MSTP110 | NADRIN | RICH1 | WBP15 | Rho GTPase activating protein 17 | Reconstituted Complex | BioGRID | 11431473 |
| AXL | JTK11 | UFO | AXL receptor tyrosine kinase | Biochemical Activity | BioGRID | 9178760 |
| AXL | JTK11 | UFO | AXL receptor tyrosine kinase | Axl interacts with an unspecified isoform of p85-alpha. | BIND | 12470648 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD,BioGRID | 10799562 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Affinity Capture-Western | BioGRID | 9461587 |9918857 |
| CBLB | DKFZp686J10223 | DKFZp779A0729 | DKFZp779F1443 | FLJ36865 | FLJ41152 | Nbla00127 | RNF56 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | Affinity Capture-Western | BioGRID | 9614102 |
| CD19 | B4 | MGC12802 | CD19 molecule | - | HPRD | 7528218 |
| CD22 | FLJ22814 | MGC130020 | SIGLEC-2 | SIGLEC2 | CD22 molecule | - | HPRD | 8647200 |
| CD28 | MGC138290 | Tp44 | CD28 molecule | - | HPRD,BioGRID | 7737275 |8621607 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | - | HPRD | 10339567 |
| CD3E | FLJ18683 | T3E | TCRE | CD3e molecule, epsilon (CD3-TCR complex) | - | HPRD,BioGRID | 9312149 |11689561 |11855827 |
| CD4 | CD4mut | CD4 molecule | - | HPRD | 8246987 |
| CD5 | LEU1 | T1 | CD5 molecule | - | HPRD,BioGRID | 9079809 |
| CD7 | GP40 | LEU-9 | TP41 | Tp40 | CD7 molecule | - | HPRD,BioGRID | 8918688 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | - | HPRD,BioGRID | 7629060 |8034624 |9381982 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | - | HPRD | 9381982 |
| CHRNA7 | CHRNA7-2 | NACHRA7 | cholinergic receptor, nicotinic, alpha 7 | - | HPRD | 11278378 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | Affinity Capture-Western | BioGRID | 9461587 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | - | HPRD,BioGRID | 8947469 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | CSF1R (c-FMS) interacts with PIK3R1 (PI3'-k).his interaction was modeled on a demonstrated interaction between CSF1R and PIK3R1 from unspecified species. | BIND | 15735664 |
| CSF2RA | CD116 | CDw116 | CSF2R | CSF2RAX | CSF2RAY | CSF2RX | CSF2RY | GM-CSF-R-alpha | GMCSFR | GMR | MGC3848 | MGC4838 | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | - | HPRD,BioGRID | 12538575 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | - | HPRD | 7807015 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Reconstituted Complex | BioGRID | 10477752 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 11716761 |
| DOK1 | MGC117395 | MGC138860 | P62DOK | docking protein 1, 62kDa (downstream of tyrosine kinase 1) | Reconstituted Complex | BioGRID | 11071635 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD | 8662998 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR interacts with p85. This interaction was modeled on a demonstrated interaction between human EGFR and bovine p85. | BIND | 1372092 |
| EPHA2 | ECK | EPH receptor A2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7982920 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD,BioGRID | 7559499 |
| ERAS | HRAS2 | HRASP | MGC126691 | MGC126693 | ES cell expressed Ras | - | HPRD | 12774123 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 1351056 |
| ERBB3 | ErbB-3 | HER3 | LCCS2 | MDA-BF-1 | MGC88033 | c-erbB-3 | c-erbB3 | erbB3-S | p180-ErbB3 | p45-sErbB3 | p85-sErbB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | - | HPRD,BioGRID | 10383151 |11546794 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 11689445 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD,BioGRID | 10377409 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD | 11741599 |
| FCGR2A | CD32 | CD32A | CDw32 | FCG2 | FCGR2 | FCGR2A1 | FcGR | IGFR2 | MGC23887 | MGC30032 | Fc fragment of IgG, low affinity IIa, receptor (CD32) | - | HPRD,BioGRID | 8631888 |
| FES | FPS | feline sarcoma oncogene | - | HPRD,BioGRID | 8916957 |
| FLT1 | FLT | VEGFR1 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | Flt-1 interacts with p85. | BIND | 7657594 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD | 8394019 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | Reconstituted Complex | BioGRID | 8294442 |
| GAB1 | - | GRB2-associated binding protein 1 | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8596638 |9891995 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | Affinity Capture-Western Two-hybrid | BioGRID | 11334882 |11782427 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD | 10068651 |
| GAB3 | - | GRB2-associated binding protein 3 | - | HPRD | 11739737 |
| GHR | GHBP | growth hormone receptor | - | HPRD,BioGRID | 9632636 |
| GNAQ | G-ALPHA-q | GAQ | guanine nucleotide binding protein (G protein), q polypeptide | Affinity Capture-Western | BioGRID | 12704201 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | p85 interacts with Grb2. | BIND | 8662998 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 7642542 |7737969 |7759531 |
| HCST | DAP10 | DKFZP586C1522 | KAP10 | PIK3AP | hematopoietic cell signal transducer | Protein-peptide | BioGRID | 10426994 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | - | HPRD | 10970851 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | p85 interacts with hnRNPK. This interaction was modeled on a demonstrated interaction between p85 from an unspecified source and human hnRNPK. | BIND | 8810341 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD,BioGRID | 10212255 |
| IFNAR1 | AVP | IFN-alpha-REC | IFNAR | IFNBR | IFRC | interferon (alpha, beta and omega) receptor 1 | - | HPRD,BioGRID | 10542297 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 8697095 |
| IL13 | ALRH | BHR1 | IL-13 | MGC116786 | MGC116788 | MGC116789 | P600 | interleukin 13 | - | HPRD | 9139718 |
| IL1R1 | CD121A | D2S1473 | IL-1R-alpha | IL1R | IL1RA | P80 | interleukin 1 receptor, type I | - | HPRD,BioGRID | 9360994 |10373529 |
| IL1RAP | C3orf13 | FLJ37788 | IL-1RAcP | IL1R3 | interleukin 1 receptor accessory protein | - | HPRD,BioGRID | 10373529 |
| IL2RB | CD122 | P70-75 | interleukin 2 receptor, beta | - | HPRD,BioGRID | 9774657 |
| IL6ST | CD130 | CDw130 | GP130 | GP130-RAPS | IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | Affinity Capture-Western | BioGRID | 10579793 |
| INPP5D | MGC104855 | MGC142140 | MGC142142 | SHIP | SHIP1 | SIP-145 | hp51CN | inositol polyphosphate-5-phosphatase, 145kDa | Affinity Capture-Western | BioGRID | 9918857 |
| INSR | CD220 | HHF5 | insulin receptor | INSR (IR) interacts with PIK3R1. This interaction was modeled on a demonstrated interaction between INSR and PIK3R1 from unspecified species. | BIND | 15735664 |
| INSR | CD220 | HHF5 | insulin receptor | The insulin receptor (IR) interacts with p85-alpha. | BIND | 8603569 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | Affinity Capture-Western Far Western | BioGRID | 1381348 |12107746 |12173038 |12730242 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD | 1381348 |12173038 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD | 12220227 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD,BioGRID | 9852124 |12107746 |
| IRS4 | IRS-4 | PY160 | insulin receptor substrate 4 | - | HPRD | 11912194 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9774657 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 8702385 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10467411 |11604231 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | Sam68 interacts with the SH2 domains of PI3K p85 alpha. This interaction was modelled based on a demonstrated interaction between two rat proteins. | BIND | 7537265 |10437794 |11604231 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | Kit interacts with p85-alpha. This interaction was modeled on a demonstrated interaction between mouse proteins | BIND | 10022833 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 7509796 |
| LAT | LAT1 | pp36 | linker for activation of T cells | Affinity Capture-Western Reconstituted Complex | BioGRID | 11368773 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD | 7504174 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | Reconstituted Complex | BioGRID | 8294442 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | Affinity Capture-Western Two-hybrid | BioGRID | 15388330 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | MET (c-Met) interacts with PIK3R1. This interaction was modeled on a demonstrated interaction between human c-Met and PIK3R1 from an unspecified species. | BIND | 15735664 |
| MME | CALLA | CD10 | DKFZp686O16152 | MGC126681 | MGC126707 | NEP | membrane metallo-endopeptidase | - | HPRD | 12529960 |
| MST1R | CD136 | CDw136 | PTK8 | RON | macrophage stimulating 1 receptor (c-met-related tyrosine kinase) | - | HPRD | 8918464 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | - | HPRD,BioGRID | 9892650 |
| NTRK2 | GP145-TrkB | TRKB | neurotrophic tyrosine kinase, receptor, type 2 | Two-hybrid | BioGRID | 12074588 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | in vivo | BioGRID | 10697503 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Beta-PDGFR interacts with p85. This interaction was modeled on a demonstrated interaction between human beta-PDGFR and rat or bovine p85. | BIND | 1372092 |7689724 |7876130 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Phosphorylated beta-PDGFR interacts with p85-alpha. This interaction was modeled on a demonstrated interaction between human beta-PDGFR and bovine p85-alpha. | BIND | 8388538 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | Affinity Capture-Western | BioGRID | 9774384 |
| PIK3CA | MGC142161 | MGC142163 | PI3K | p110-alpha | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Reconstituted Complex | BioGRID | 7929193 |
| PIK3CB | DKFZp779K1237 | MGC133043 | PI3K | PI3KCB | PI3Kbeta | PIK3C1 | p110-BETA | phosphoinositide-3-kinase, catalytic, beta polypeptide | - | HPRD,BioGRID | 8139559 |10358930 |
| PIK3CD | p110D | phosphoinositide-3-kinase, catalytic, delta polypeptide | - | HPRD,BioGRID | 9113989 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Reconstituted Complex | BioGRID | 8294442 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 7537275 |8649427 |10806474 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD | 10797305 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Affinity Capture-Western | BioGRID | 9918857 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | Two-hybrid | BioGRID | 11160222 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD | 11337495 |
| PXN | FLJ16691 | paxillin | Co-purification | BioGRID | 8641358 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | - | HPRD | 7629060 |8034624 |12086876 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | - | HPRD | 12086876 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD | 1336372 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Rb interacts with p55alpha | BIND | 12588990 |
| RET | CDHF12 | HSCR1 | MEN2A | MEN2B | MTC1 | PTC | RET-ELE1 | RET51 | ret proto-oncogene | - | HPRD | 10652352 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD,BioGRID | 12454018 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | - | HPRD,BioGRID | 11850823 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | - | HPRD,BioGRID | 10921882 |
| SHB | RP11-3J10.8 | bA3J10.2 | Src homology 2 domain containing adaptor protein B | Shb interacts with p85-alpha PI3 kinase. This interaction was modelled on a demonstrated interaction between human Shb and bovine p85-alpha PI3 kinase. | BIND | 7537362 |
| SHB | RP11-3J10.8 | bA3J10.2 | Src homology 2 domain containing adaptor protein B | - | HPRD,BioGRID | 7537362 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Reconstituted Complex | BioGRID | 7537361 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | p85 interacts with Shc. | BIND | 8662998 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD | 8662998 |
| SLC9A2 | NHE2 | solute carrier family 9 (sodium/hydrogen exchanger), member 2 | - | HPRD | 10187839 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 12210743 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 10487518 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD,BioGRID | 10899172 |
| TEK | CD202B | TIE-2 | TIE2 | VMCM | VMCM1 | TEK tyrosine kinase, endothelial | - | HPRD | 10521483 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD,BioGRID | 9435577 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | Affinity Capture-Western | BioGRID | 9435577 |
| TIE1 | JTK14 | TIE | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | - | HPRD | 11865050 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | Co-purification | BioGRID | 8641358 |
| TOM1L1 | OK/KNS-CL.3 | SRCASM | target of myb1 (chicken)-like 1 | - | HPRD | 11711534 |
| TRAT1 | HSPC062 | TCRIM | TRIM | T cell receptor associated transmembrane adaptor 1 | - | HPRD | 10723796 |
| TUBA1B | K-ALPHA-1 | tubulin, alpha 1b | - | HPRD,BioGRID | 7592789 |
| TUBG1 | TUBG | TUBGCP1 | tubulin, gamma 1 | - | HPRD | 7592789 |
| TYRO3 | BYK | Brt | Dtk | FLJ16467 | RSE | Sky | Tif | TYRO3 protein tyrosine kinase | - | HPRD,BioGRID | 10627473 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD | 7528218 |9891995 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Affinity Capture-Western Reconstituted Complex | BioGRID | 9162069 |9891995 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD | 11094073 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | Affinity Capture-Western Reconstituted Complex | BioGRID | 8805332 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD | 10358777 |
| WBP11 | DKFZp779M1063 | NPWBP | SIPP1 | WW domain binding protein 11 | - | HPRD | 11375989 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 3731 | 3737 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-128 | 481 | 487 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-129-5p | 3922 | 3928 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC | ||||
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-15/16/195/424/497 | 3732 | 3739 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-153 | 800 | 807 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 1327 | 1333 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-182 | 3590 | 3596 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-185 | 68 | 74 | 1A | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-21 | 1103 | 1109 | m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-218 | 3555 | 3562 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-29 | 327 | 333 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-3p | 1639 | 1645 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-320 | 350 | 356 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-33 | 283 | 289 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-361 | 1100 | 1106 | 1A | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-376 | 996 | 1002 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-448 | 801 | 807 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-450 | 286 | 292 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-486 | 952 | 959 | 1A,m8 | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-493-5p | 703 | 709 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-503 | 3733 | 3739 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-96 | 3590 | 3596 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.